Recreating the U.S. River Conditions animations in R
Use R to generate a video and gif similar to the Vizlab animation series, U.S. River Conditions.
What's on this page
For the last few years, we have released quarterly animations of streamflow conditions at all active USGS streamflow sites . These visualizations use daily streamflow measurements pulled from the USGS National Water Information System (NWIS) to show how streamflow changes throughout the year and highlight the reasons behind some hydrologic patterns. Here, I walk through the steps to recreate a similar version in R.
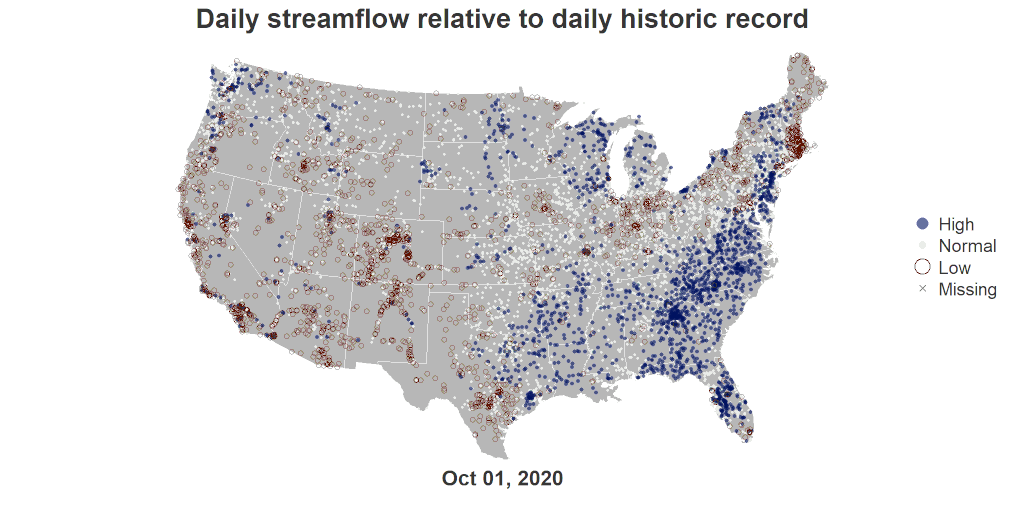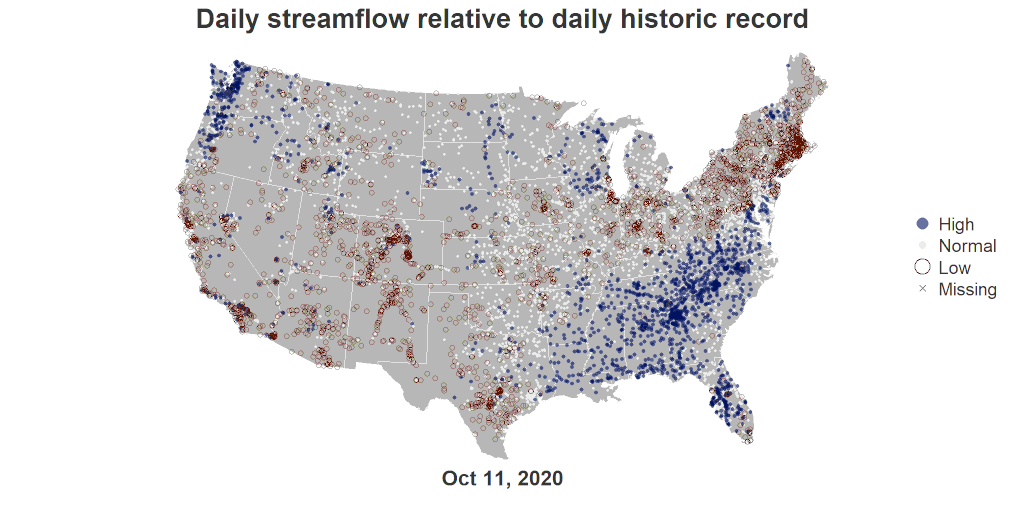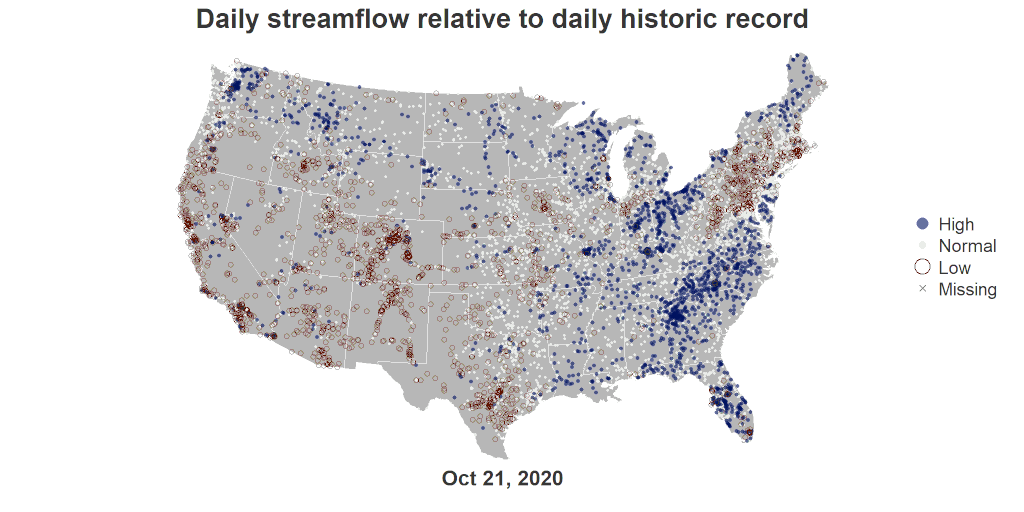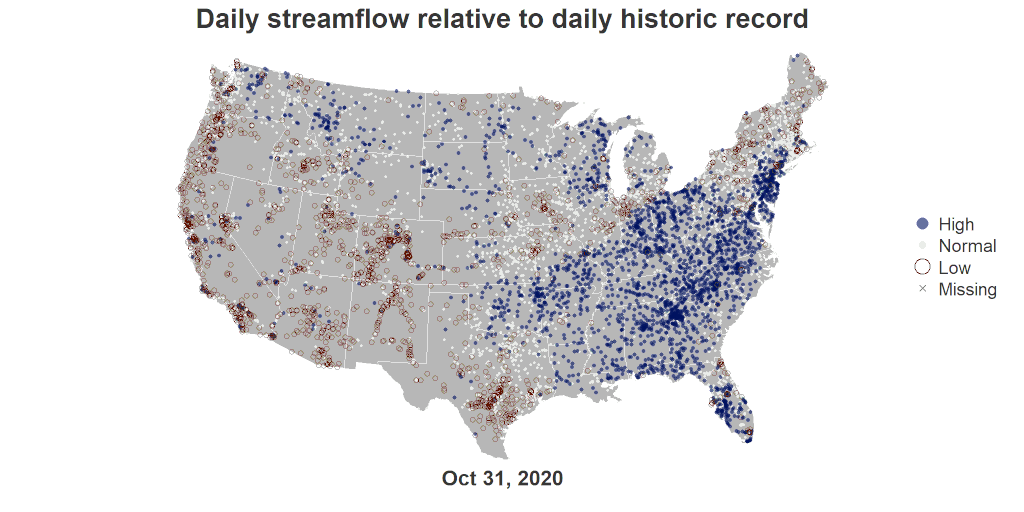
The workflow to recreate the U.S. River Conditions animations can be divided into these key sections:
- Workflow setup
- Fetch data
- Prepare data for mapping
- Create animation frames
- Combine frames into a gif
- Combine frames into a video
- Optimize your animations
Background on the animation series
The U.S. River Conditions animation series (visit the full portfolio of USGS Vizlab products to see examples) was inspired by the USGS streamflow conditions website, WaterWatch . WaterWatch depicts national streamflow conditions by coloring each gage’s dot by how it’s daily streamflow value relates to that day’s record of streamflow values. Hydrologists and other water data experts use this nuanced metric to inform daily decisions.
In contrast to WaterWatch, the U.S. River Conditions animation series was intended to target a new audience of users who may be unfamiliar with the USGS streamgaging network. To make the content more accessible to non-technical audiences, we deviated from WaterWatch by 1) adding context to the data by including callout text, 2) building a file type that can be directly embedded on social media platforms (e.g Twitter, Facebook, Instagram), and 3) using streamflow metrics that were not as complex. To make the streamflow metrics less complex, the animation colors each gage’s dot by how it’s daily streamflow value relates to its entire record of streamflow values (not the individual day’s record as WaterWatch does). This change helps reinforce knowledge that lots of people already have about seasonal patterns (e.g. wet springs and dry summers) and builds on that existing framework to introduce streamflow data.
We rebuild the U.S. River Conditions animation every three months to summarize the last quarter’s streamflow
events to the public. To be able to do this efficiently and programmatically, the entire visualization is
coded in R. The only manual effort required is to update the timeframe of interest and
configure the text for hydrologic events. You can see the code for the visualization
here on GitHub
.
The visualization pipeline is completely reproducible, but does use a custom, internal
pipelining tool built off of the R package, remake (very powerful, but
almost like learning a new language). To make the processing more transparent and accessible,
I created this blog to demonstrate the main workflow for how we create the U.S. River Conditions
animations in R (independent of the pipeline tool). The workflow below will
not recreate them exactly, but it should give you the tools to make similar
video and gif animations from R.
Setup your workflow
This code is set up to operate based on all CONUS states. You should be able to change the state(s), projection, and date range to quickly generate something relevant to your area!
library(dataRetrieval)
library(tidyverse) # includes dplyr, tidyr
library(maps)
library(sf)
# Viz dates
viz_date_start <- as.Date("2020-10-01")
viz_date_end <- as.Date("2020-10-31")
# Currently set to CONUS (to list just a few, do something like `c("MN", "WI", "IA", "IL")`)
viz_states <- c("AL", "AZ", "AR", "CA", "CO", "CT", "DE", "FL", "GA",
"ID", "IL", "IN", "IA", "KS", "KY", "LA", "ME", "MD", "MA",
"MI", "MN", "MS", "MO", "MT", "NE", "NV", "NH", "NJ", "NM", "NY",
"NC", "ND", "OH", "OK", "OR", "PA", "RI", "SC", "SD", "TN", "TX",
"UT", "VT", "VA", "WA", "WV", "WI", "WY")
# Albers Equal Area, which is good for full CONUS
viz_proj <- "+proj=laea +lat_0=45 +lon_0=-100 +x_0=0 +y_0=0 +a=6370997 +b=6370997 +units=m +no_defs"
# Configs that could be changed ...
param_cd <- "00060" # Parameter code for streamflow . See `dataRetrieval::parameterCdFile` for more.
service_cd <- "dv" # means "daily value". See `https://waterservices.usgs.gov/rest/` for more info.
Fetch your data
To fetch data for this example, will be using the USGS R
package, dataRetrieval. To learn more about how to use these functions and discover other
capabilities of the package, see Laura DeCicco’s blog post: dataRetrieval Tutorial - Using R to Discover Data
. You can also
explore our online, self-paced course here
.
Find the site numbers
Find the gages that have data for your states and time period.
# Can only query one state at a time, so need to loop through.
# For one month of CONUS data, this took ~ 1 minute.
sites_available <- data.frame() # This will be table of site numbers and their locations.
for(s in viz_states) {
# Some states might not return anything, so use `tryCatch` to allow
# the `for` loop to keep going.
message(sprintf("Trying %s ...", s))
sites_available <- tryCatch(
whatNWISsites(
stateCd = s,
startDate = viz_date_start,
endDate = viz_date_end,
parameterCd = param_cd,
service = service_cd) %>%
select(site_no, station_nm, dec_lat_va, dec_long_va),
error = function(e) return()
) %>% bind_rows(sites_available)
}
head(sites_available)
site_no station_nm dec_lat_va dec_long_va
1 13027500 SALT RIVER ABOVE RESERVOIR, NEAR ETNA, WY 43.07972 -111.0372
2 10020100 BEAR RIVER ABOVE RESERVOIR, NEAR WOODRUFF, UT 41.43439 -111.0177
3 10020300 BEAR RIVER BELOW RESERVOIR, NEAR WOODRUFF, UT 41.50550 -111.0146
4 13025500 CROW CREEK NEAR FAIRVIEW, WY 42.67493 -111.0077
5 10028500 BEAR RIVER BELOW PIXLEY DAM, NEAR COKEVILLE, WY 41.93883 -110.9855
6 13023000 GREYS RIVER ABOVE RESERVOIR, NEAR ALPINE, WY 43.14306 -110.9769
Next, get the statistics data
The biggest processing hurdle for the U.S. River Conditions animation is
to fetch and process EVERY streamflow data point in the entire NWIS
database in order to calculate historic statistics. We have a separate
pipeline to pull the data (see
national-flow-observations
)
and steps in the U.S. River Conditions code to calculate the percentiles
(see this
script
).
However, for the purposes of this workflow, we will use the stat service (beta)
from NWIS
so that we don’t need to pull down all of the historic data. This means
that the final viz will show the values relative to that day’s historic
values (as WaterWatch does); however, using the stat service will
greatly simplify the processing part of this example so we can get to the
animation!
For this map, we will create a scale of 3 categories from low (less than 25th percentile), normal (between 25th and 75th percentile), and high (greater than 75th percentile). So, let’s fetch those stats for the sites we found in the last code block. You can visit the stats service documentation for more on what stats are available to download and this website to learn about percentiles in streamflow .
# For one month of CONUS, this took ~ 15 min
# Can only pull stats for 10 sites at a time, so loop through chunks of 10
# sites at a time and combine into one big dataset at the end!
sites <- unique(sites_available$site_no)
# Divide request into chunks of 10
req_bks <- seq(1, length(sites), by=10)
viz_stats <- data.frame()
for(chk_start in req_bks) {
chk_end <- ifelse(chk_start + 9 < length(sites), chk_start + 9, length(sites))
# Note that each call throws warning about BETA service :)
message(sprintf("Pulling down stats for %s:%s", chk_start, chk_end))
viz_stats <- tryCatch(readNWISstat(
siteNumbers = sites[chk_start:chk_end],
parameterCd = param_cd,
statType = c("P25", "P75")
) %>% select(site_no, month_nu, day_nu, p25_va, p75_va),
error = function(e) return()
) %>% bind_rows(viz_stats)
}
head(viz_stats)
site_no month_nu day_nu p25_va p75_va
1 02339495 1 1 40 209
2 02339495 1 2 59 248
3 02339495 1 3 53 317
4 02339495 1 4 48 464
5 02339495 1 5 41 270
6 02339495 1 6 39 283
Now get the daily values
Now that we have our sites and their statistics, we can pull the data for the timeframe of our visualization. These are the values that will be compared with the site statistics to determine if our gage is experiencing low, normal, or high streamflow conditions.
# For all of CONUS, there are 8500+ sites.
# Just in case there is an issue, use a loop to cycle through 100
# at a time and save the output temporarily, then combine after.
# For one month of CONUS data, this took ~ 10 minutes
max_n <- 100
sites <- unique(sites_available$site_no)
sites_chunk <- seq(1, length(sites), by=max_n)
site_data_fns <- c()
tmp <- tempdir()
for(chk_start in sites_chunk) {
chk_end <- ifelse(chk_start + (max_n-1) < length(sites),
chk_start + (max_n-1), length(sites))
site_data_fn_i <- sprintf(
"%s/site_data_%s_%s_chk_%s_%s.rds", tmp,
format(viz_date_start, "%Y_%m_%d"), format(viz_date_end, "%Y_%m_%d"),
# Add leading zeroes in front of numbers so they remain alphanumerically ordered
sprintf("%02d", chk_start),
sprintf("%02d", chk_end))
site_data_fns <- c(site_data_fns, site_data_fn_i)
if(file.exists(site_data_fn_i)) {
message(sprintf("ALREADY fetched sites %s:%s, skipping ...", chk_start, chk_end))
next
} else {
site_data_df_i <- readNWISdata(
siteNumbers = sites_available$site_no[chk_start:chk_end],
startDate = viz_date_start,
endDate = viz_date_end,
parameterCd = param_cd,
service = service_cd
) %>% renameNWISColumns() %>%
select(site_no, dateTime, Flow)
saveRDS(site_data_df_i, site_data_fn_i)
message(sprintf("Fetched sites %s:%s out of %s", chk_start, chk_end, length(sites)))
}
}
# Read and combine all of them
viz_daily_values <- lapply(site_data_fns, readRDS) %>% bind_rows()
nrow(viz_daily_values) # ~250,000 obs for one month of CONUS data
head(viz_daily_values)
site_no dateTime Flow
1 05344490 2020-10-01 3000
2 05344490 2020-10-02 3010
3 05344490 2020-10-03 2890
4 05344490 2020-10-04 2660
5 05344490 2020-10-05 2360
6 05344490 2020-10-06 2600
Process and prepare your data for mapping
Categorize daily values by comparing to the statistics
We now have all of the data! Next, we can categorize each day’s data into low, normal, or high based on the statistics. The category is what we will use to determine how to style the points for each frame of the animation.
# To join with the stats data, we need to first break the dates into
# month day columns
viz_daily_stats <- viz_daily_values %>%
mutate(month_nu = as.numeric(format(dateTime, "%m")),
day_nu = as.numeric(format(dateTime, "%d"))) %>%
# Join the statistics data and automatically match on month_nu and day_nu
left_join(viz_stats) %>%
# Compare the daily flow value to that sites statistic to
# create a column that says whether the value is low, normal, or high
mutate(viz_category = ifelse(
Flow <= p25_va, "Low",
ifelse(Flow > p25_va & Flow <= p75_va, "Normal",
ifelse(Flow > p75_va, "High",
NA))))
head(viz_daily_stats)
site_no dateTime Flow month_nu day_nu p25_va p75_va viz_category
1 05344490 2020-10-01 3000 10 1 3210 6540 Low
2 05344490 2020-10-02 3010 10 2 3150 6430 Low
3 05344490 2020-10-03 2890 10 3 3080 6430 Low
4 05344490 2020-10-04 2660 10 4 3360 6970 Low
5 05344490 2020-10-05 2360 10 5 3410 8180 Low
6 05344490 2020-10-06 2600 10 6 3330 9860 Low
Setup visualization configs
I like to separate visual configurations, such as frame dimensions and symbol colors/shapes, so that it is easy to find them when I want to make adjustments or reuse the code for another project.
# Viz frame height and width. These are double the size the end video &
# gifs will be. See why in the "Optimizing for various platforms" section
viz_width <- 2048 # in pixels
viz_height <- 1024 # in pixels
# Style configs for plotting (all are listed in the same order as the categories)
viz_categories <- c("High", "Normal", "Low", "Missing")
category_col <- c("#00126099", "#EAEDE9FF", "#601200FF", "#7f7f7fFF") # Colors (hex + alpha code)
category_pch <- c(16, 19, 1, 4) # Point types in order of categories
category_cex <- c(1.5, 1, 2, 0.75) # Point sizes in order of categories
category_lwd <- c(NA, NA, 1, NA) # Circle outline width in order of categories
Get data ready for mapping
We are going to need to do two last things to get our data ready
to visualize. First, add columns to specify the style of each point called viz_pt_color,
viz_pt_type, viz_pt_size, and viz_pt_outline. These will be passed in to our
plotting step so that each point has the appropriate style applied. Second, join in the latitude
and longitude columns (dec_lat_va, dec_long_va) and then convert to a
spatial data frame.
# Created a function to use that picks out the correct config based on
# category name. Made it a fxn since I needed to do it for colors,
# point types, and point sizes. This function assumes that all of the
# config vectors are in the order High, Normal, Low, and Missing.
pick_config <- function(viz_category, config) {
ifelse(
viz_category == "Low", config[3],
ifelse(viz_category == "Normal", config[2],
ifelse(viz_category == "High", config[1],
config[4]))) # NA color
}
# Add style information as 3 new columns
viz_data_ready <- viz_daily_stats %>%
mutate(viz_pt_color = pick_config(viz_category, category_col),
viz_pt_type = pick_config(viz_category, category_pch),
viz_pt_size = pick_config(viz_category, category_cex),
viz_pt_outline = pick_config(viz_category, category_lwd)) %>%
# Simplify columns
select(site_no, dateTime, viz_category, viz_pt_color,
viz_pt_type, viz_pt_size, viz_pt_outline) %>%
left_join(sites_available) %>%
# Arrange so that NAs are plot first, then normal values on top of those, then
# high values, and then low so that viewer can see the most points possible.
arrange(dateTime, factor(viz_category, levels = c(NA, "Normal", "High", "Low")))
# Convert to `sf` object for mapping
viz_data_sf <- viz_data_ready %>%
st_as_sf(coords = c("dec_long_va", "dec_lat_va"), crs = "+proj=longlat +datum=WGS84") %>%
st_transform(viz_proj)
head(viz_data_sf)
Simple feature collection with 6 features and 8 fields
geometry type: POINT
dimension: XY
bbox: xmin: -856104.2 ymin: -129028.2 xmax: 567670.3 ymax: 17517.14
CRS: +proj=laea +lat_0=45 +lon_0=-100 +x_0=0 +y_0=0 +a=6370997 +b=6370997 +units=m +no_defs
site_no dateTime viz_category viz_pt_color viz_pt_type viz_pt_size viz_pt_outline
1 05344500 2020-10-01 Normal #EAEDE9FF 19 1 NA
2 06036805 2020-10-01 Normal #EAEDE9FF 19 1 NA
3 06037100 2020-10-01 Normal #EAEDE9FF 19 1 NA
4 06186500 2020-10-01 Normal #EAEDE9FF 19 1 NA
5 06218500 2020-10-01 Normal #EAEDE9FF 19 1 NA
6 06220800 2020-10-01 Normal #EAEDE9FF 19 1 NA
station_nm geometry
1 MISSISSIPPI RIVER AT PRESCOTT, WI POINT (567670.3 -3040.174)
2 Firehole River at Old Faithful, YNP POINT (-855744.5 -2936.8)
3 Gibbon River at Madison Jct, YNP POINT (-856104.2 17517.14)
4 Yellowstone River at Yellowstone Lk Outlet YNP POINT (-819525.6 4401.4)
5 WIND RIVER NEAR DUBOIS, WY POINT (-783904.9 -111020.9)
6 WIND RIVER ABOVE RED CREEK, NEAR DUBOIS, WY POINT (-761597.7 -129028.2)
Get background map data ready
Using the states the user picked, create an sf object to use as the
map background.
# Convert state codes into lowercase state names, which `maps` needs
viz_state_names <- tolower(stateCdLookup(viz_states, "fullName"))
state_sf <- st_as_sf(maps::map('state', viz_state_names, fill = TRUE, plot = FALSE)) %>%
st_transform(viz_proj)
head(state_sf)
Simple feature collection with 6 features and 1 field
geometry type: MULTIPOLYGON
dimension: XY
bbox: xmin: -2029655 ymin: -1556239 xmax: 2295848 ymax: 68939.91
CRS: +proj=laea +lat_0=45 +lon_0=-100 +x_0=0 +y_0=0 +a=6370997 +b=6370997 +units=m +no_defs
ID geom
1 alabama MULTIPOLYGON (((1207268 -15...
2 arizona MULTIPOLYGON (((-1329818 -9...
3 arkansas MULTIPOLYGON (((557029.8 -1...
4 california MULTIPOLYGON (((-1633107 -1...
5 colorado MULTIPOLYGON (((-175185.3 -...
6 connecticut MULTIPOLYGON (((2141452 238...
Build the frames
Test by making a single frame
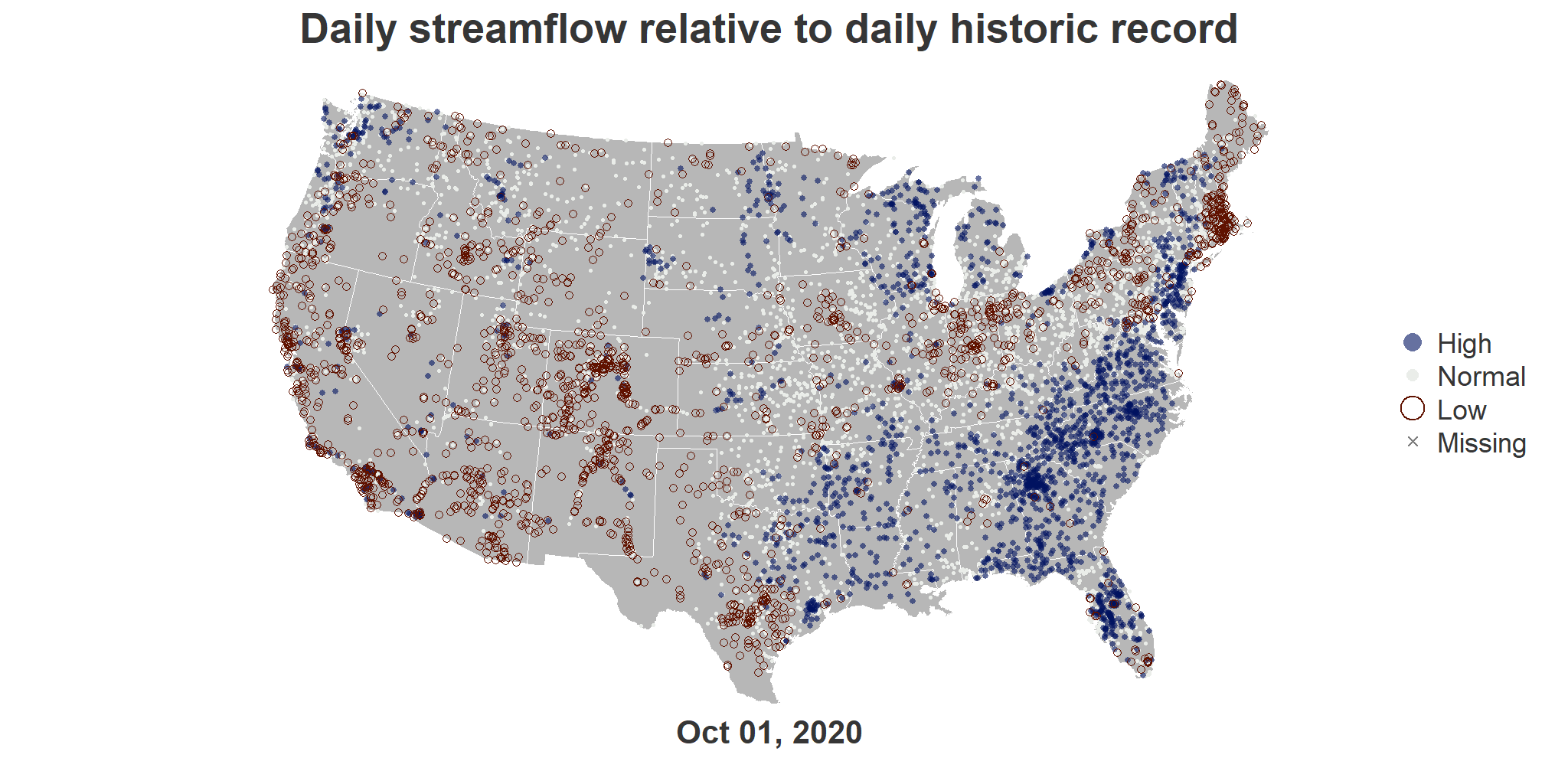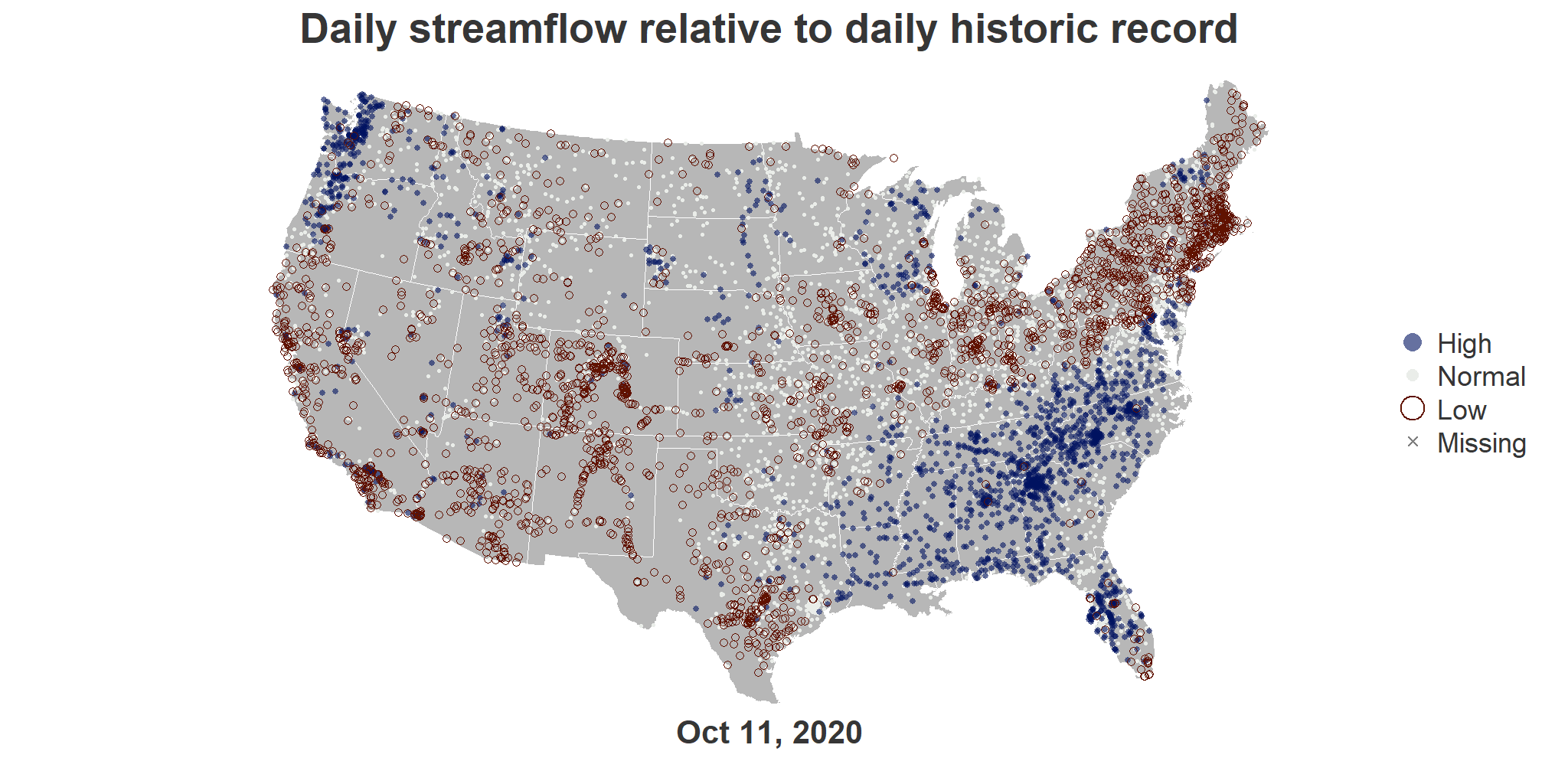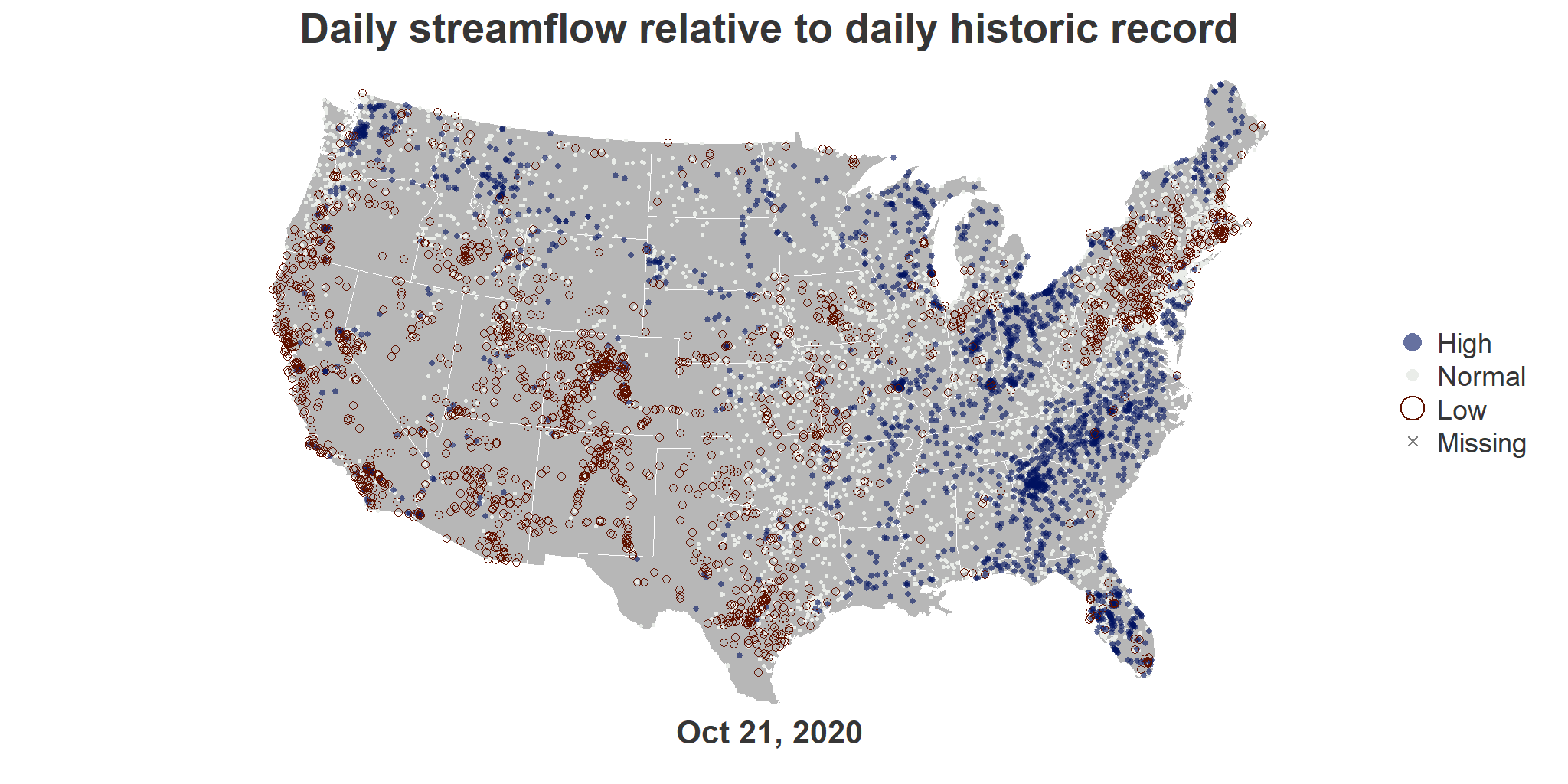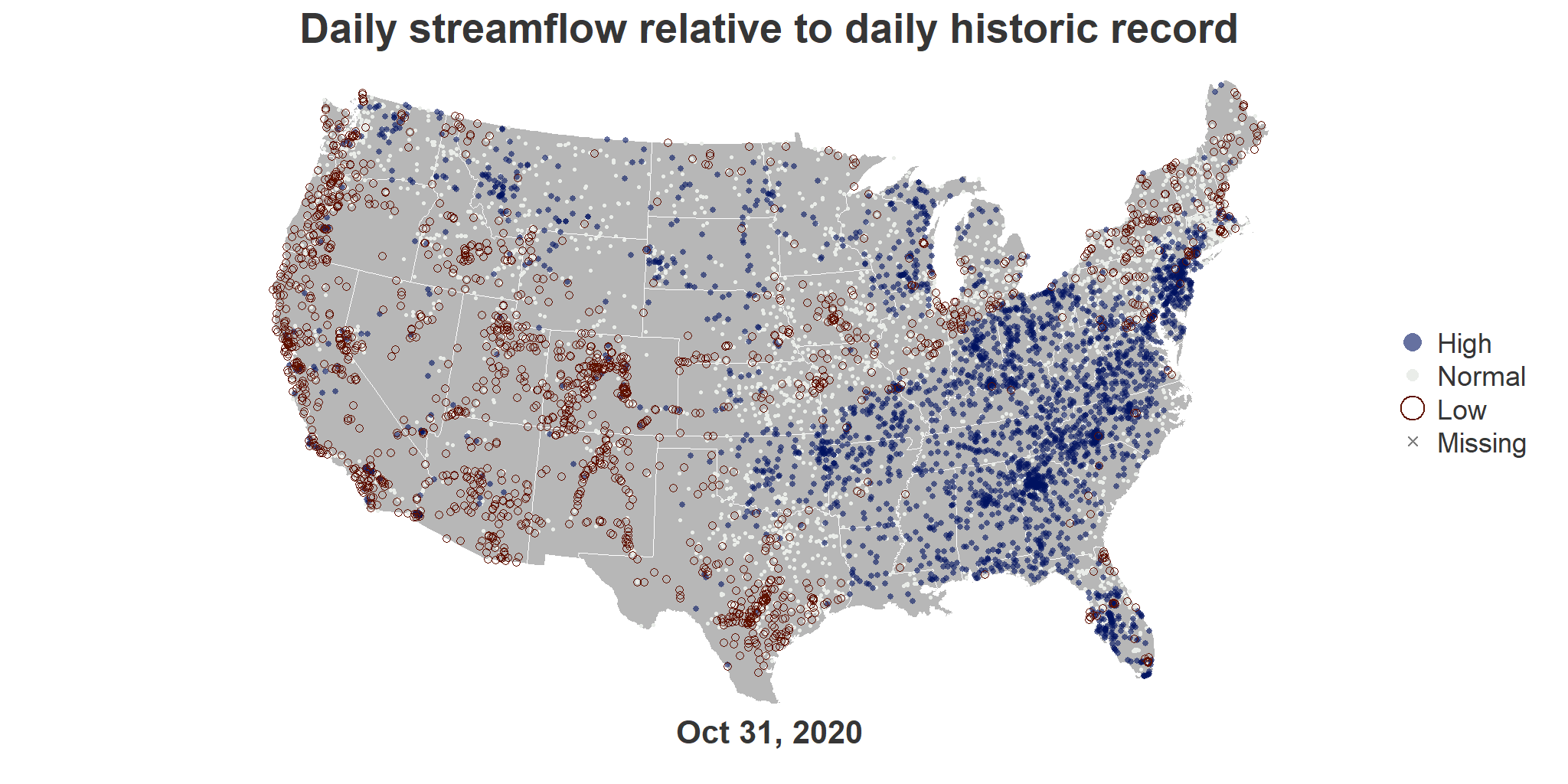
To create each frame, we use base R plotting functions. In the end, there will be one PNG file per day. Pick the first day in the date sequence to test out a single frame and adjust style, as needed, with this single frame before moving on to the full build.
png("test_single_day.png", width = viz_width, height = viz_height)
# Prep plotting area (no outer margins, small space at top/bottom for title, white background)
par(omi = c(0,0,0,0), mai = c(0.5,0,0.5,0.25), bg = "white")
# Add the background map
plot(st_geometry(state_sf), col = "#b7b7b7", border = "white")
# Add gages for the first day in the data set
viz_data_sf_1 <- viz_data_sf %>% filter(dateTime == viz_date_start)
plot(st_geometry(viz_data_sf_1), add=TRUE, pch = viz_data_sf_1$viz_pt_type,
cex = viz_data_sf_1$viz_pt_size, col = viz_data_sf_1$viz_pt_color,
lwd = viz_data_sf_1$viz_pt_outline)
# Now add on a title, the date, and a legend
text_color <- "#363636"
title("Daily streamflow relative to daily historic record", cex.main = 4.5, col.main = text_color)
mtext(format(viz_date_start, "%b %d, %Y"), side = 1, line = 0.5, cex = 3.5, font = 2, col = text_color)
legend("right", legend = viz_categories, col = category_col, pch = category_pch,
pt.cex = category_cex*3, pt.lwd = category_lwd*2, bty = "n", cex = 3, text.col = text_color)
dev.off()
You should now have a PNG file in your working directory called test_single_day.png. When you open this file, you should see the following image (unless you made customizations along the way). Continue to iterate here before moving on if you would like to edit the style because it is easiest to do with just a single frame.
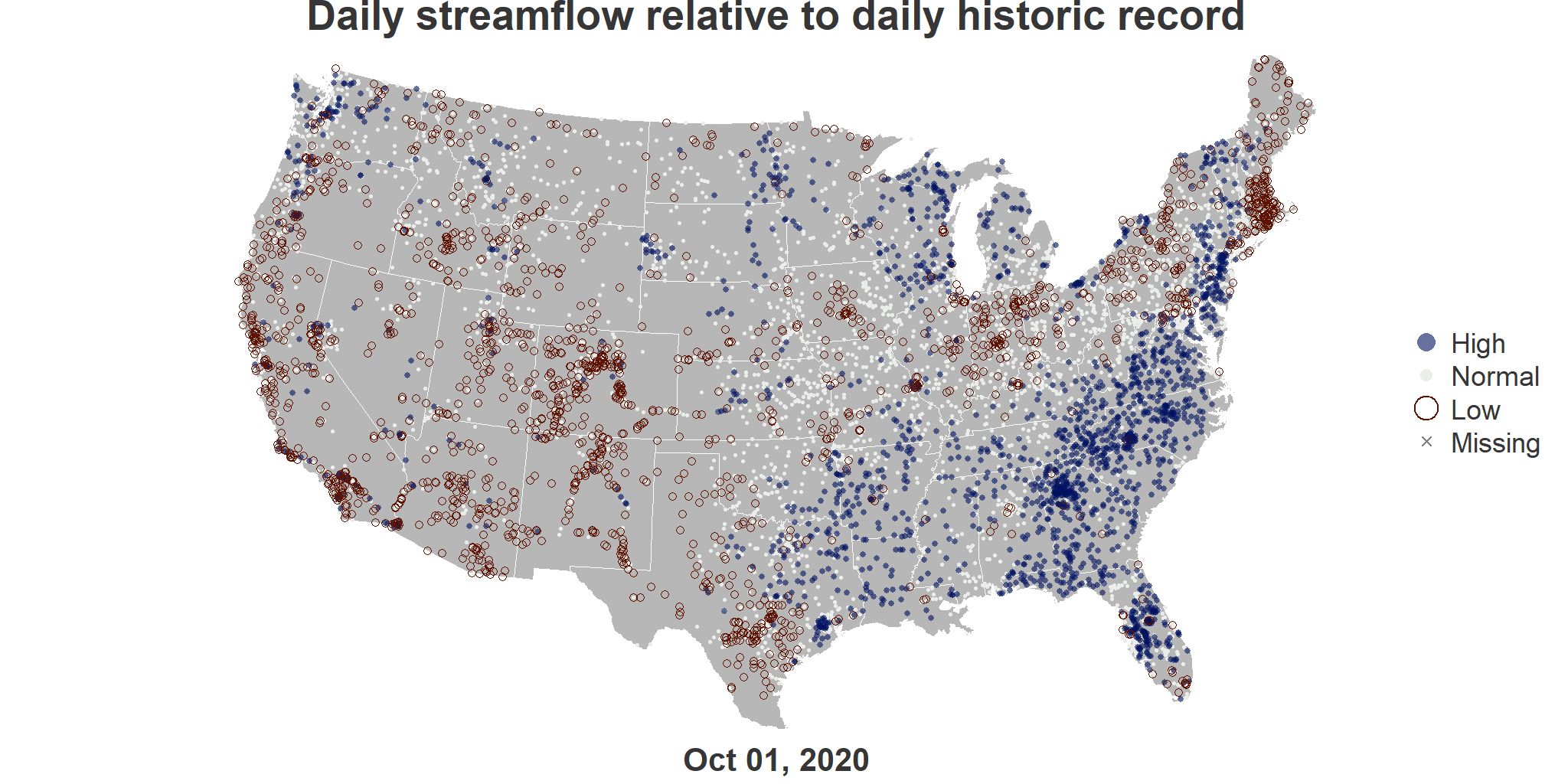
Create all animation frames
Now that we are satisified with how our single frame looks, you can move on to building one frame per day.
# This takes ~2 minutes to create a frame per day for a month of data
frame_dir <- "frame_tmp" # Save frames to a folder
frame_configs <- data.frame(date = unique(viz_data_sf$dateTime)) %>%
# Each day gets it's own file
mutate(name = sprintf("%s/frame_%s.png", frame_dir, date),
frame_num = row_number())
if(!dir.exists(frame_dir)) dir.create(frame_dir)
# Loop through dates
for(i in frame_configs$frame_num) {
this_date <- frame_configs$date[i]
# Filter gage data to just this day:
data_this_date <- viz_data_sf %>% filter(dateTime == this_date)
png(frame_configs$name[i], width = viz_width, height = viz_height) # Open file
# Copied plotting code from previous step
# Prep plotting area (no outer margins, small space at top/bottom for title, white background)
par(omi = c(0,0,0,0), mai = c(1,0,1,0.5), bg = "white")
# Add the background map
plot(st_geometry(state_sf), col = "#b7b7b7", border = "white")
# Add gages for this day
plot(st_geometry(data_this_date), add=TRUE, pch = data_this_date$viz_pt_type,
cex = data_this_date$viz_pt_size, col = data_this_date$viz_pt_color,
lwd = data_this_date$viz_pt_outline)
# Add the title, date, and legend
text_color <- "#363636"
title("Daily streamflow relative to daily historic record",
cex.main = 4.5, col.main = text_color)
mtext(format(this_date, "%b %d, %Y"), side = 1, line = 0.5,
cex = 3.5, font = 2, col = text_color)
legend("right", legend = viz_categories, col = category_col,
pch = category_pch, pt.cex = category_cex*3,
pt.lwd = category_lwd*2, bty = "n", cex = 3,
text.col = text_color)
dev.off() # Close file
}
# You can check that all the files were created by running
all(file.exists(frame_configs$name))
Combine frames into animations
Now that you have all of your frames for the animation ready, we can stitch them together into a GIF or a video!
GIF
To create GIFs, I use a software called ImageMagick. It is not an R package,
but we can run its commands by wrapping them in the system function in R. A colleage
recently told me about the
R package magick
,
which is a wrapper for ImageMagick and would allow you to avoid the system function.
It sounds great but I just learned about it and have not had time to practice it yet.
I might return to this post in the future with an update using magick once I learn it.
For these commands to work, you will first need to install the
ImageMagick library. Go to
imagemagick.org
to do so.
Once installed, make sure you restart R if you had it open before
installing.
# GIF configs that you set
frames <- frame_configs$name # Assumes all are in the same directory
gif_length <- 10 # Num seconds to make the gif
gif_file_out <- sprintf("animation_%s_%s.gif",
format(viz_date_start, "%Y_%m_%d"),
format(viz_date_end, "%Y_%m_%d"))
# ImageMagick needs a temporary directory while it is creating the GIF to
# store things. Make sure it has one. I am making this one, inside of
# the directory where our frames exist.
tmp_dir <- sprintf("%s/magick", dirname(frames[1]))
if(!dir.exists(tmp_dir)) dir.create(tmp_dir)
# Calculate GIF delay based on desired length & frames
# See https://legacy.imagemagick.org/discourse-server/viewtopic.php?t=14739
fps <- length(frames) / gif_length # Note that some platforms have specific fps requirements
gif_delay <- round(100 / fps) # 100 is an ImageMagick default (see that link above)
# Create a single string of all the frames going to be stitched together
# to pass to ImageMagick commans.
frame_str <- paste(frames, collapse = " ")
# Create the ImageMagick command to convert the files into a GIF
magick_command <- sprintf(
'convert -define registry:temporary-path=%s -limit memory 24GiB -delay %s -loop 0 %s %s',
tmp_dir, gif_delay, frame_str, gif_file_out)
# If you are on a Windows machine, you will need to have `magick` added before the command
if(Sys.info()[['sysname']] == "Windows") {
magick_command <- sprintf('magick %s', magick_command)
}
# Now actually run the command and create the GIF
system(magick_command)
After running that code, you should now have a GIF file called animation_2020_10_01_2020_10_31.gif
in your working directory! An optional step is to use an additional non-R library called gifsicle
to simplify the GIF. It will lower the final size of your GIF (which
can get quite large if you have a lot of frames). You will need to
install gifsicle
before you can run
this code.
# Simplify the gif with gifsicle - cuts size by about 2/3
gifsicle_command <- sprintf('gifsicle -b -O3 -d %s --colors 256 %s',
gif_delay, gif_file_out)
system(gifsicle_command)
Video
Videos are created in a similar way to GIFs, but I use a tool called
ffmpeg. There are a lot of configuration options (I am still
learning about how to use these). The options used in the code below are
what we have found results in the best video. You can see all of the
options and download the tool from
ffmpeg.org
.
Note: This tool requires an extra step if you are on a Windows machine because it cannot use your filenames outright (unless numbered from 1:n) . I have included the extra step to add the filenames to a text file and use that in the video creation command .
# Video configs that you set
frames <- frame_configs$name # Assumes all are in the same directory
video_length <- 30 # in seconds
video_file_out <- sprintf("animation_%s_%s.mp4",
format(viz_date_start, "%Y_%m_%d"),
format(viz_date_end, "%Y_%m_%d"))
# Calculate frame rate based on desired length & num frames
# If r < 1, duplicates some frames; if r > 1, drops some frames
# Round up to nearest 0.25 (don't need to be more specific than that)
r <- video_length/length(frames)
r <- ceiling(r / 0.25) * 0.25
# Put all frame filenames into a single text file & their
# durations in the line right below (use same duration for
# each in this example).
files_to_cat_fn <- "frames_to_video.txt" # Will delete at end
writeLines(sprintf("file '%s'\nduration %s", frames, r), files_to_cat_fn)
# Construct command, see more options at https://www.ffmpeg.org/ffmpeg.html
# `-y` allows you to overwrite the out file
# `-f` concat is needed to tell ffmpeg we are using a text file of images to use
# `-i %s` will list the text file for input files to stitch together
# `-pix_fmt yuv420p` is the pixel format being used. This works well with libx264
# `-vcodec libx264` choose the library that will be used to compress the video
# `-crf 27`, option for controling quality. See https://slhck.info/video/2017/02/24/crf-guide.html
ffmpeg_command <- sprintf(
"ffmpeg -y -f concat -i %s -pix_fmt yuv420p -vcodec libx264 -crf 27 %s",
files_to_cat_fn, video_file_out)
system(ffmpeg_command) # Run command
This takes a few seconds longer than the GIF creation steps and will print a whole
lot of messages out to your console as it runs. Once it is complete, you will see
a new mp4 file in your working directory called, animation_2020_10_01_2020_10_31.mp4 and
it should look a little something like this one:
This process of creating individual frames and them stitching together into a video or GIF
will work for any set of images you have. To use this code with other frames (if you didn’t
create the ones above), just change out the first line of the GIF or
video code chunks that define the object, frames, which contains the filepaths of the images
you want to convert to an animation.
Optimizing for various platforms
Image quality
For Windows users, the quality of saved PNG images is pretty low and results in pixelated videos. We were able to overcome this by setting the width and height to double the desired size and later downscaling the video (see this GitHub issue to read about our discovery process for fixing Windows pixelation). If you change the size of the frames you are saving after you iterate on your frame design, you will need to adjust your point and text sizes to fit the new image dimensions and then recreate the animation files.
When creating the PNG files earlier, I used a width and height double the size of what I wanted in the end, so now all I have to do is downscale. If you followed this approach, you can downscale your video to half the size by running the following:
# For GIFs
original_gif_file <- gif_file_out # The file with the dimensions double what you need
out_gif_file <- gsub(".gif", "_downscaled.gif", gif_file_out) # The final file that has been downscale
system(sprintf("gifsicle --scale 0.5 -i %s > %s", original_gif_file, out_gif_file))
# For videos
original_video_file <- video_file_out # The file with the dimensions double what you need
out_video_file <- gsub(".mp4", "_downscaled.mp4", video_file_out) # The final file that has been downscale
system(sprintf("ffmpeg -i %s -vf scale=iw/2:-1 %s", original_video_file, out_video_file))
You will now have two new animation files, one GIF and one video, that have _downscaled
appended to their names. They should appear to be about half of the size of the originals
(my GIF went from 817 KB to 441 KB and my video went from 2 MB to 900 KB).
Platform specs
Different social media platforms have different requirements for your content. The most important item to check before planning your animation are the dimensions and aspect ratio you will need. Those can change the way that you need to layout your animation (for example, Instagram videos are often square or portrait but rarely landscape). If I want to put this on multiple platforms, you will likely need to generate multiple animations. I always have three different versions, where each is optimized for the platform on which it will be released. When I do this, I make sure to write functions to share as much of the code as possible but deviate as necessary.
As you plan your animation, consider the following list of technical specifications that platforms may require. To find technical specs by platform, you can use your favorite web search! Try to look at the most recent list available because the specs can change. So far, I have had great success with this updating article from Sprout Social .
- maximum file size
- maximum video length
- maximum/minimum height and width
- specific aspect ratio
- framerate
Other animation options
In the world of R, there are lots of other animation options. There is
an R package called magick that wraps the ImageMagick library we used earlier (I only recently
discovered this and may try to update this blog in the future using
that package instead of the system calls). If you are
a ggplot2 user, there is a package called gganimate that will create
animations without the need for any of the additional tools I listed
here. I have struggled with using that package in the past (though to be
fair since I had been using this method, I didn’t try very hard to
learn) and found it to be very slow compared to the multiframe plus ImageMagick
or FFMPEG method.
There are also non-R tools you can use to stitch together animation frames, such as Photoshop. In addition to not having a Photoshop license, I am a huge R nerd, so the more I can stay in R, the better :)
That should give you some foundations and tools to build frame-based animations in R. Good luck and let us know how it goes @USGS_DataSci !
Categories:
Related Posts
Duplicating Quarto elements with code templates to reduce copy and paste errors
May 20, 2025
Introduction
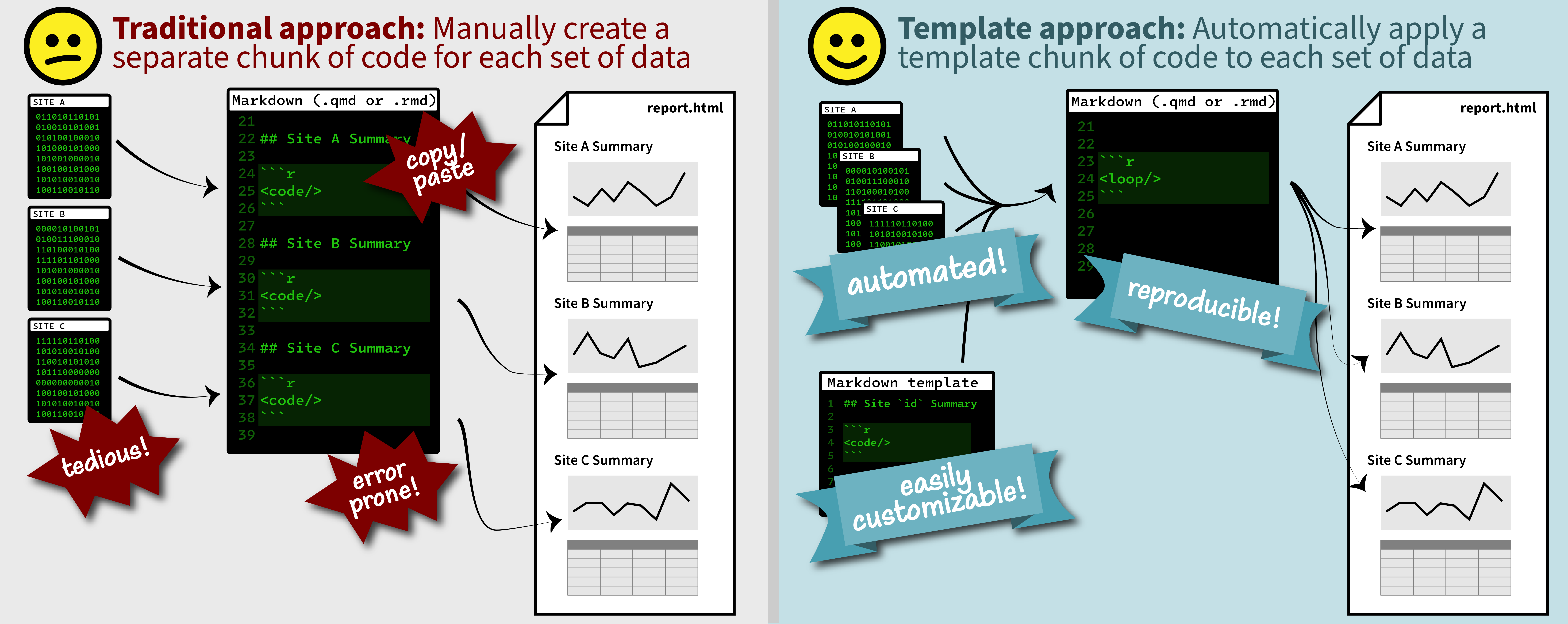
It is a common situation in data science and analysis: We want to create a series of figures, tables, or summary statistics for a set of data. Maybe we’re studying different species of irises or penguins or the current flow conditions for various streamgages (the example used below), and we want a different summary figure or table for each. One common approach is to write code for one set of the data, such as setting up the graphing parameters in a
ggplotdata visualization or calculating a series of statistics for a single species/streamgage. Then, once happy with that, copying and pasting the code for each entity, modifying the code slightly for each iteration.Jazz up your ggplots!
July 21, 2023
At Vizlab , we make fun and accessible data visualizations to communicate USGS research and data. Upon taking a closer look at some of our visualizations, you may think:
Beyond Basic R - Mapping
August 16, 2018
Introduction
There are many different R packages for dealing with spatial data. The main distinctions between them involve the types of data they work with — raster or vector — and the sophistication of the analyses they can do. Raster data can be thought of as pixels, similar to an image, while vector data consists of points, lines, or polygons. Spatial data manipulation can be quite complex, but creating some basic plots can be done with just a few commands. In this post, we will show simple examples of raster and vector spatial data for plotting a watershed and gage locations, and link to some other more complex examples.
Exploring ggplot2 boxplots - Defining limits and adjusting style
August 10, 2018
Boxplots are often used to show data distributions, and
ggplot2is often used to visualize data. A question that comes up is what exactly do the box plots represent? Theggplot2box plots follow standard Tukey representations, and there are many references of this online and in standard statistical text books. The base R function to calculate the box plot limits isboxplot.stats. The help file for this function is very informative, but it’s often non-R users asking what exactly the plot means. Therefore, this post breaks down the calculations into (hopefully!) easy-to-follow chunks of code for you to make your own box plot legend if necessary. Some additional goals here are to create boxplots that come close to USGS style. Features in this post take advantage of enhancements toggplot2in version 3.0.0 or later.Beyond Basic R - Plotting with ggplot2 and Multiple Plots in One Figure
August 9, 2018
R can create almost any plot imaginable and as with most things in R if you don’t know where to start, try Google. The Introduction to R curriculum summarizes some of the most used plots, but cannot begin to expose people to the breadth of plot options that exist.There are existing resources that are great references for plotting in R:

