Large sample pulls using dataRetrieval
Large pull for all total nitrogen data for streams in the contiguous United States with sites that have at least 40 measurements between 1995 and 2020.
dataRetrieval is an R package that provides user-friendly access to
water data from either the Water Quality Portal (WQP) or the National
Water Information Service (NWIS). dataRetrieval makes it easier for a
user to download data from the WQP or NWIS and convert it into usable
formats.
This blog will walk through an example of a large data pull to increase the reusability, reproducibility, and efficiency of WQP data workflows. The script-based workflow uses the WQP summary service
to limit the amount downloaded to only relevant data, which can save a lot of time and ultimately reduce the
complexity of subsequent data processing. This post is an alternative method to the targets workflow presented in Large Data Pulls from WQP - A Pipeline-Based Approach
.
Large data example
This blog sets up a scenario to look at all the total nitrogen data measured in streams within the contiguous United States where sites have at least 40 measurements between 1995 and 2020.
Data download
First, set up a loop to find out which sites have the required data, then get only the data that is relevant.
There are several ways to break up the geographic part of the query,
such as bounding box, state, county, or hydrologic unit code (HUC).
Depending on the density of the data, it may be ideal to adjust how to
loop through the geographic query. Sometimes running a single
dataRetrieval call comes back with a “timeout” error. Other times,
requests for data spans more than traditional geographic filters of HUC,
state, or county. In these cases, it may be necessary to break up the
dataRetrieval call into smaller subsets, and then bind those subsets
together separately. This blog will discuss one strategy for breaking up
a WQP request.
dataRetrieval includes a data frame “stateCd” that has all of the
states and territories that could be queried by either NWIS or WQP. In
this example, only the lower 48 states along with Washington, D.C.are
considered.
Use the readWQPsummary function, which is a very useful function that
returns information about all the data that is available for a
particular query. The initial query is asking what nitrogen data for
streams is available in a particular state. The returned data shows
how many nitrogen samples are available at each site for each year.
Then, using filtering and summaries will figure out exactly which sites
meet the set up scenairo’s needs.
The readWQPdata function is used to download the actual relevant data.
This example saves the data using the saveRDS function for each
individual state. This ensures higher likelihood of successful
completion of the query. For example, if a failure occurs during the
download and the loops don’t finish. In that case, the states that
successfully downloaded the data are skipped, and only re-run the states
that didn’t work. Saving as an “RDS” file also has the benefits of
retaining all the attributes of the data. Notice another feature of this
loop is using tryCatch for each of the dataRetrieval calls. This
allows the loop to continue even if one of the states failed for some
reason.
# Load packages
library(dataRetrieval)
library(tidyverse)
state_cd_cont <- stateCd[c(2,4:12,14:52),] # state code information for the 48 conterminous United States plus DC
rownames(state_cd_cont) <- seq(length=nrow(state_cd_cont)) # reset row sequence
for(i in seq_len(nrow(state_cd_cont))){
state_cd <- state_cd_cont$STATE[i]
state_nm <- state_cd_cont$STUSAB[i]
message("Getting: ", state_nm)
df_summary <- tryCatch({
readWQPsummary(statecode = state_cd,
CharacteristicName = "Nitrogen",
siteType = "Stream")
},
error=function(cond) {
message(paste("No data in:", state_nm))
break()
})
sites <- df_summary |>
filter(YearSummarized >= 1995,
YearSummarized <= 2020) |>
group_by(MonitoringLocationIdentifier, MonitoringLocationName) |>
summarise(start_year = min(YearSummarized, na.rm = TRUE),
end_year = max(YearSummarized, na.rm = TRUE),
count_activity = sum(ActivityCount, na.rm = TRUE),
count_result = sum(ResultCount, na.rm = TRUE)) |>
ungroup() |>
filter(count_activity >= 40)
if(nrow(sites) > 0){
df_state <- tryCatch({
readWQPdata(siteid = sites$MonitoringLocationIdentifier,
CharacteristicName = "Nitrogen",
startDateLo = "1995-01-01",
startDateHi = "2020-12-31",
sampleMedia = "Water",
ignore_attributes = FALSE)
# turning off attributes speeds things
# up, but then you'll need to do the site info later.
# But...you'll have filtered the number of sites down.
# If you think that'll be a lot of sites filtered out,
# set ignore_attributes to TRUE
},
error=function(cond) {
message(paste("No data in:", state_nm))
})
}
if(nrow(df_state) > 0){
# I would write the data here, just in case:
saveRDS(df_state, file = paste0(state_nm, "_raw_data.rds"))
# You could slim it down here,
# but if later you decide you wanted something you filtered
# out here, you'd need to remember to get the data again
# For example:
df_state2 <- df_state |>
filter(ResultSampleFractionText == "Total") |>
group_by(MonitoringLocationIdentifier) |>
mutate(count = n()) |>
filter(count > 40) |>
ungroup()
} else {
message("No data in:", state_nm)
}
}
The following can be included in the loop above, but saving it for later allows for more flexibility with the raw data (e.g., leaving data in or filtering data out).
Although creating an empty data frame and filling the data in later would be the most efficient way to go, binding the rows is flexible and easy to conceptualize. For this data download scenario, there wasn’t a huge bottleneck by using the “dyplr::bind_rows”, but that could be a place to reconsider if the next section seems to be taking too long, in which creating the empty data frame may be the considered solution.
The next loop shown below opens each file, pulls out the data we need
for analysis, and binds up each state into one large data frame. Notice
“ResultMeasureValue” changes into a character vector. By default,
dataRetrieval will try to convert that column into numeric. Sometimes
however, that can’t be done because there are actual character values in
the results. Therefore, to retain all of that information, be sure each
state’s “ResultMeasureValue” column is a character (more information on
this is below).
all_nitrogen <- data.frame()
# for(state in stateCd$STUSAB){
for(state in state_cd_cont$STUSAB){
state_df <- tryCatch({
readRDS(paste0(state, "_raw_data.rds"))
})
if(nrow(state_df) > 0){
df_slim <- state_df |>
mutate(ResultMeasureValue = as.character(ResultMeasureValue),
`DetectionQuantitationLimitMeasure.MeasureValue` = as.character(`DetectionQuantitationLimitMeasure.MeasureValue`)) |>
filter(ActivityMediaSubdivisionName %in%
c("Surface Water")|
is.na(ActivityMediaSubdivisionName),
ResultSampleFractionText %in% c("Total")) |>
left_join(attr(state_df, "siteInfo") |>
select(MonitoringLocationIdentifier,
dec_lat_va, dec_lon_va,
MonitoringLocationName,
MonitoringLocationTypeName,
HUCEightDigitCode),
by = "MonitoringLocationIdentifier")
all_nitrogen <- bind_rows(all_nitrogen, df_slim)
}
}
Results can be provided in a non-numeric format (e.g. “no data”, “bdl”, “<0.1”), which is why it is important to keep the result in character form:
unique(all_nitrogen$ResultMeasureValue[which(!is.na(all_nitrogen$ResultMeasureValue) &
is.na(as.numeric(all_nitrogen$ResultMeasureValue)))])
## [1] "N/A" "no data" "bdl"
## [4] "BDL" "below detection limits" "..198"
## [7] "*Non-detect" "0,8" "<0.50000000"
## [10] ">0.00000000" "<1.00000000" "<0.05000000"
## [13] "<0.00000000"
Explore the output
Visually view the data available
To identify the geospatial footprint of sites that meet the above data
download criteria, create a map showing all site locations. Here, the
maps package is used to create the base map of the conterminous United
States, fills in state outlines, and draws the site locations using the
latitude and longitude coordinates from the data download.
# Load packages
library(maps)
# create the base map so that map extents are correct
map("usa")
# add state outlines
map("state", add=TRUE)
# draw the site locations onto th map
points(all_nitrogen$dec_lon_va, all_nitrogen$dec_lat_va, col="red", pch=20)
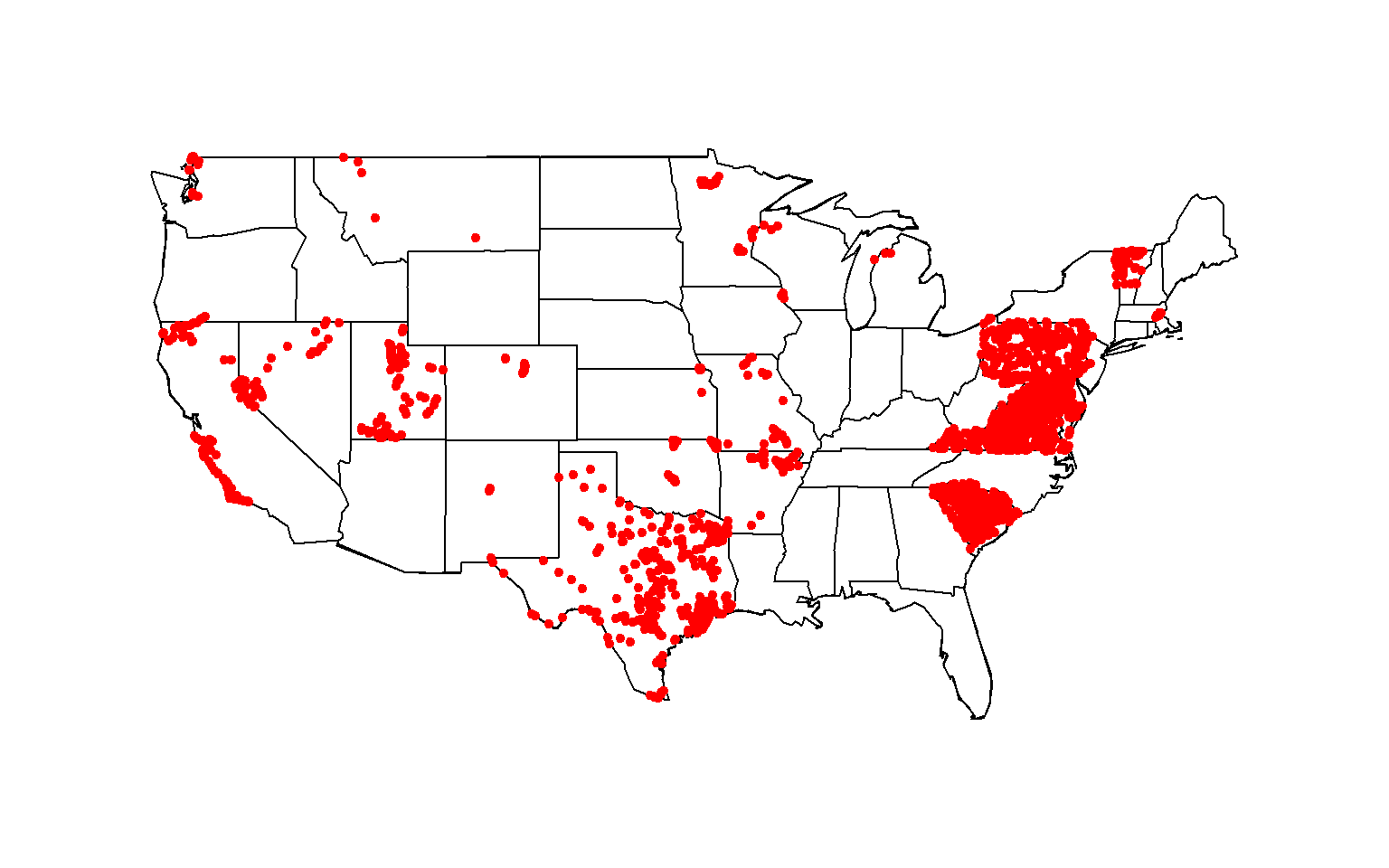
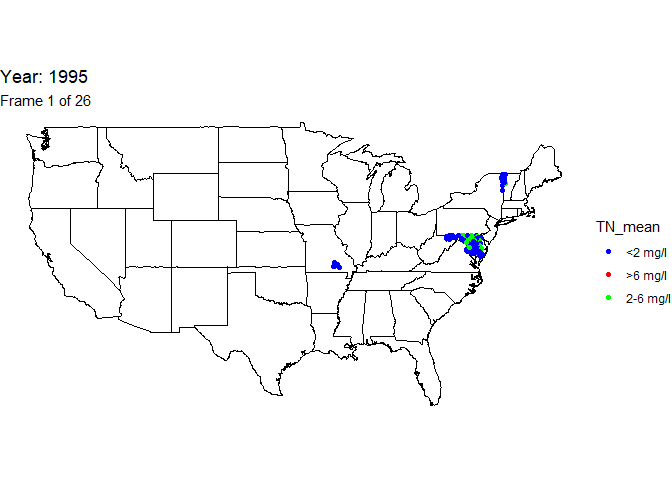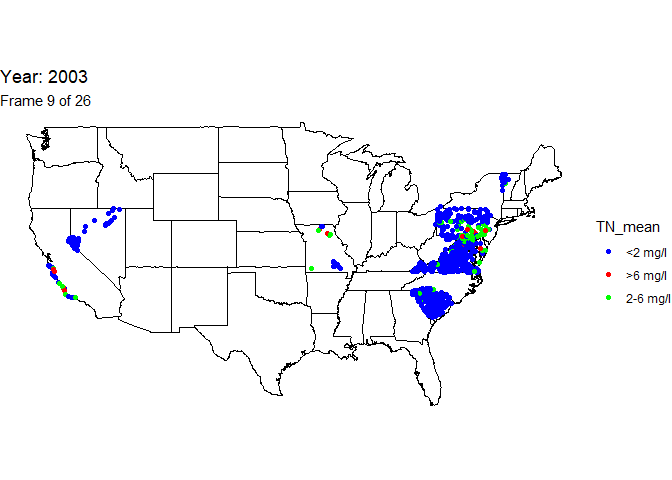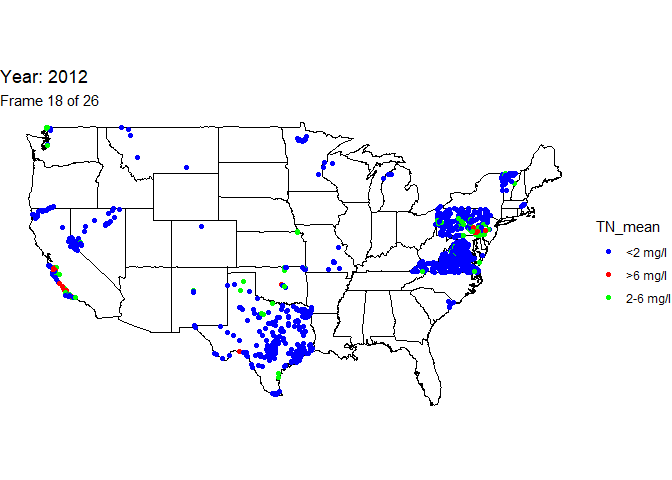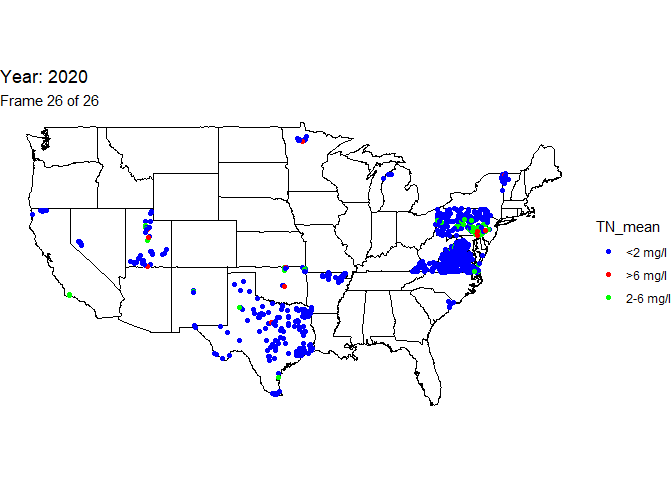
Total nitrogen data available at stream site locationed throughout the contiguous United States, 1995-2020
Data exploration and data filtering
Depending on the purpose of the data, non-numeric result values may or may not be useful. Although total nitrogen standards differ between states, suppose your purpose is to compare total nitrogen data among sites between 1995 and 2020 to the U.S. Environmental Protection Agency (EPA) total nitrogen acceptable range of 2-6 milligrams per liter (mg/l; U.S. Environmental Protection Agency, 2013). With this defined purpose, you would be looking at field data with known concentrations. In order to compare the data to the EPA acceptable range, the data would need to be in mg/l.
Reference: U.S. Environmental Protection Agency, 2013, Total Nitrogen - Revised: EPA 090-R-090-20A.
First, explore the unique sample types:
# determine sample types
unique(all_nitrogen$ActivityTypeCode)
## [1] "Sample-Routine"
## [2] "Quality Control Sample-Field Blank"
## [3] "Sample-Field Split"
## [4] "Field Msr/Obs"
## [5] "Sample-Other"
## [6] "Quality Control Sample-Field Replicate"
## [7] "Quality Control Sample-Blind Duplicate"
## [8] "Quality Control Sample-Equipment Blank"
## [9] "Sample-Field Subsample"
## [10] "Quality Control Sample-Lab Duplicate"
## [11] "Sample-Integrated Vertical Profile"
## [12] "Quality Control Sample-Inter-lab Split"
## [13] "Quality Control Field Sample Equipment Rinsate Blank"
## [14] "Sample-Composite Without Parents"
## [15] "Field Msr/Obs-Habitat Assessment"
## [16] "Sample-Integrated Cross-Sectional Profile"
## [17] "Sample-Integrated Horizontal Profile"
## [18] "Sample-Integrated Time Series"
Second, explore the reported result units:
# determine units used to report total nitrogen
unique(all_nitrogen$ResultMeasure.MeasureUnitCode)
## [1] "mg/l" "ppm" NA "ug/l" "MPN/100ml" "ug"
In the above two snippet of code and output, you see there are field and quality assurance data reported in a variety of units.
Third, use piping (%>%) and the filter function from the dplyr
package to filter out quality assurance data and then filter result
units in weight per unit volume of water.
# filter out quality control data by using the "filter" function, and then use the "filter" function again to keep only results reported in wight per unit volume of water.
all_nitrogen <- all_nitrogen %>%
filter(!grepl("Quality Control", ActivityTypeCode)) %>%
filter(ResultMeasure.MeasureUnitCode=="mg/l" | ResultMeasure.MeasureUnitCode=="ppm" | ResultMeasure.MeasureUnitCode=="ug/l")
In the above snippet of code, the pipe operator (%>%) takes the output of the first function in front of it and passes it to the second function, which allows you to express a sequence of multiple operations.
Now, explore the non-numeric result values. Here, create a new data
frame “nonnum” using piping and the filter function again to filter
non-numeric result values that are also not reported as NA.
nonnum <- all_nitrogen %>%
filter(ResultMeasureValue %>%
as.numeric() %>%
is.na())
Viewing the “nonnum” data frame, some result values were reported as censored with a “<” or “>” character or simply appear to have been entered with typos (i.e., “..198 and 0,8), while other result values are reported as censored below the detection limit (i.e.,”bdl”, “BDL”, “below detection limits”) or no data reported (i.e., “N/A”, “no data”, or NA). With that, the user would have to make decisions on how to use these data.
Next, explore the non-numeric result values. Create a new data frame named “nitrogen” with only pertinent columns to make the data easier to work with. Then, create a new column of result values named “Result”. The purpose of creating a new results column is to retain the original results data while still being able work with the data (unit conversions, non-numeric to numeric, etc.).
# create a new data frame with only pertinent columns from the existing all_nitrogen data frame
nitrogen <- all_nitrogen[,c("OrganizationIdentifier", "OrganizationFormalName", "ActivityIdentifier", "ActivityTypeCode", "ActivityStartDate", "ActivityCommentText", "ResultDetectionConditionText", "CharacteristicName", "ResultSampleFractionText", "ResultMeasureValue", "ResultMeasure.MeasureUnitCode", "MeasureQualifierCode", "ResultStatusIdentifier", "ResultValueTypeName", "ResultCommentText", "ResultLaboratoryCommentText", "DetectionQuantitationLimitTypeName", "DetectionQuantitationLimitMeasure.MeasureValue", "DetectionQuantitationLimitMeasure.MeasureUnitCode", "dec_lat_va", "dec_lon_va", "MonitoringLocationName", "MonitoringLocationIdentifier", "MonitoringLocationTypeName")]
# create new Result column using the original result value column
nitrogen$Result <- nitrogen$ResultMeasureValue
Then, for result values reported as censored or below the detection limit, create a new column named “ResultComment” to retain the censored information.
# create a new column to retain result value censoring information
nitrogen <-
nitrogen %>% mutate(ResultComment=
case_when(grepl("<", Result) ~ "<",
grepl(">", Result) ~ ">",
grepl("BDL", Result) ~ "<",
grepl("bdl", Result) ~ "<",
grepl("below detection limits", Result) ~ "<",
grepl("Not Detected", ResultDetectionConditionText) ~ "<",
grepl("Present Below Quantification Limit", ResultDetectionConditionText) ~ "<"))
Censored data are still useful for the purpose of this analysis. If a value was originally reported below a certain known threshold below the threshold of this analysis (2 mg/l), the result can still be used at that value. Now, some values that are censored do not provide the censoring limit in the results value column, but some results provide a censor threshold in the “DectectionQuantitationLimitMeasure.MeasureValue” column. In these instances, pull this information into the results column. Then, remove “<” or “>” from the “Result” column since the censoring of these data have been retained in the “ResultComment” column created in the last snippet of code.
Again, using piping, if available, fill NA result values with a censored result comment with the censoring threshold while retaining the result values that are not censored. Then, remove the “<” or “>” characters from the result column.
nitrogen <-
nitrogen %>% mutate(Result=
case_when(is.na(Result) & ResultComment=="<" ~ DetectionQuantitationLimitMeasure.MeasureValue,
!is.na(Result) ~ Result))
# remove censoring from Result column
nitrogen$Result <- gsub("<", "",
gsub(">", "", nitrogen$Result))
In the above first snippet of code, if the “Result” column is NA and the “ResultComment” column contains a “<” then, if reported, the detection limit is made the “Result” column value. While, if the “Result” column value is already provided, the result value remains the same.
Next, filter out the remaining non-numeric values that do not provide a numeric result value, including values that seem to be typos. These values should be removed because they cannot be used to compare results to the EPA acceptable range for total nitrogen (2-6 mg/l). Lastly, change the data format to numeric form. Data are converted to numeric form for unit conversions.
# filter out non-numeric values
nitrogen <-
nitrogen %>%
filter(!grepl("no data|N/A|0,8|..198|-", Result)) %>%
mutate(Result = as.numeric(Result))
There are still NA result values reported that do not indicate it is censored or is censored and does not provide a censoring threshold, so these values are not useful for this analysis and should be filtered out.
# remove NA result values
nitrogen <- nitrogen %>%
filter(!is.na(Result))
Lastly, convert all results to mg/l. Some results are already in mg/l and 1 part per million (ppm) is approximately equivalent to 1 mg/l, so these results do not need to be converted. Result values reported in micrograms per liter (ug/l) should be divided by 1000 to convert to mg/l.
# convert values to single units (milligrams per liter)
nitrogen <-
nitrogen %>% mutate(Result_mgL=
case_when(ResultMeasure.MeasureUnitCode=="ug/l" ~ Result/1000,
ResultMeasure.MeasureUnitCode=="mg/l" ~ Result,
ResultMeasure.MeasureUnitCode=="ppm" ~ Result))
Summarize data
The data shows there are multiple results per site per year. Let’s say the goal is to calculate the annual mean and median total nitrogen values for each site. First, create a new column that extracts just the year from the sample date. Then, group the data by site identifier and location (latitude and longitude) and calculate the mean and median for each year.
# pull the year from the date
nitrogen$year <- format(as.Date(nitrogen$ActivityStartDate, format="%Y-%m-%d"),"%Y")
# create new data frame of summarized result data
n_bysite <-
nitrogen %>%
group_by(MonitoringLocationIdentifier, dec_lat_va, dec_lon_va, year) %>%
summarize(mean = mean(Result_mgL),
median = median(Result_mgL))
Finally, use the total nitrogen annual mean data to create a column called “TN_mean” to indicate if the annual mean is below the EPA acceptable range (<2 mg/l), within the acceptable (2-6 mg/l), or above the acceptable (>6 mg/l).
# create a new character column with total nitrogen acceptable range designations
n_bysite <-
n_bysite %>% mutate(TN_mean=
case_when(mean<2 ~ "<2 mg/l",
mean>=2 & mean<=6 ~ "2-6 mg/l",
mean>6 ~ ">6 mg/l"))
Create animated map
An animation is a powerful visualization tool used to show how data
changes over time. Animate the data by year and result value based on
EPA acceptable standard range for total nitrogen in streams (2-6
milligrams per liter). Create this animated map using the maps,
ggplot2, gganimate, and gifski packages.
First, before plotting out data on to a map, create a data frame that
contains the base map. The maps package provides the base map boundary
and state outlines, while the map_data function from the ggplot2
package creates the data frame containing the base map data in a data
frame. The all_state variable was assigned the map boundary to all
states in the contiguous United States. In the base map data frame, the
“group” column groups the data by state (e.g., 1 = Alabama, 2 =
Arizona).
Second, plot the base map and add the data to it. Here, the base map
data that is created is used to plot the base map and the summarized
(annual mean) total nitrogen data for each site location. The total
nitrogen acceptable range data are assigned to the color parameter and
the data are grouped by year for each frame. The transition_time is
specified to use the year to move from one frame of the animation to the
next.
Lastly, run the animation using the animate function from the
gganimate package. Here the number of frame changes are equal the the
number of years in the data set.
# Load packages
library(ggplot2)
library(gganimate)
library(gifski)
# convert latitude, longitude, and year data to numeric form
n_bysite$dec_lat_va <- as.numeric(n_bysite$dec_lat_va)
n_bysite$dec_lon_va <- as.numeric(n_bysite$dec_lon_va)
n_bysite$year <- as.numeric(n_bysite$year)
# first, create the base map data frame
all_state <- "usa"
usa <- map_data("state", interior=TRUE)
base_map <- ggplot(data = usa, mapping = aes(x = long,
y = lat,
group = group)) +
geom_polygon(color = "black", fill = "white") +
coord_quickmap() +
theme_void()
# second, plot the base map and add data to it
map_with_data <- base_map +
geom_point(data = n_bysite, aes(x = dec_lon_va,
y = dec_lat_va,
color = TN_mean,
group = year,
frame = year)) +
transition_time(year) +
ggtitle('Year: {frame_time}', # add year to the title
subtitle = 'Frame {frame} of {nframes}') +
scale_colour_manual(values = c("blue", "red", "green"))
num_years <- max(n_bysite$year)-min(n_bysite$year) + 1
# lastly, run the animation
animate(map_with_data, nframes = num_years, fps = 1)
The below line of code will save the output gif:
#anim_save("TN.gif", map_with_data, nframes = num_years, fps = 1) # save
Disclaimer
Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Categories:
Related Posts
Tutorial of dataRetrieval's newest features in R
November 26, 2025
This article will describe the R-package
dataRetrieval, which simplifies the process of finding and retrieving water from the U.S. Geological Survey (USGS) and other agencies. We have recently released a new version ofdataRetrievalto work with the modernized Water Data APIs . The new version ofdataRetrievalhas several benefits for new and existing users:Hydrologic Analysis Package (HASP) Available to Users
July 26, 2022
A new R computational package was created to aggregate and plot USGS groundwater data, providing users with much of the functionality provided in Groundwater Watch and the Florida Salinity Mapper . The Hydrologic Analysis Package (HASP) can retrieve groundwater level and groundwater quality data, aggregate these data, plot them, and generate basic statistics. Documentation is available in R or online , and users can also launch a Shiny Application from within the package to generate images in an interactive user interface.
Reproducible Data Science in R: Say the quiet part out loud with assertion tests
September 2, 2025
Overview
This blog post is part of the Reprodicuble data science in R series that works up from functional programming foundations through the use of the targets R package to create efficient, reproducible data workflows.
Duplicating Quarto elements with code templates to reduce copy and paste errors
May 20, 2025
Introduction
It is a common situation in data science and analysis: We want to create a series of figures, tables, or summary statistics for a set of data. Maybe we’re studying different species of irises or penguins or the current flow conditions for various streamgages (the example used below), and we want a different summary figure or table for each. One common approach is to write code for one set of the data, such as setting up the graphing parameters in a
ggplotdata visualization or calculating a series of statistics for a single species/streamgage. Then, once happy with that, copying and pasting the code for each entity, modifying the code slightly for each iteration.Formatting guidance for USGS Samples Data Tables
May 6, 2025
Recently, changes were made to the delivery format of USGS samples data (learn more here ). In this blog, we describe the impact to users and show an example of how to use R to convert WQX-formatted water quality results data to a tabular, or “wide” view.

