The Hydro Network-Linked Data Index
Access subsets of NHDPlus data and discover sites on the river network with the hydro Network Linked Data Index. Navigation types include upstream mainstem, upstream with tributaries, downstream mainstem and downstream with diversions. Services to provide a basin boundary, linked NWIS stream gage sites, water quality data sites, and twelve digit hydrologic unit code (HUC) watershed outlets.
Introduction
updated 11-2-2020 after updates described here .
updated 9-20-2024 when the NLDI moved from labs.waterdata.usgs.gov to api.water.usgs.gov/nldi/
The Hydro Network-Linked Data Index (NLDI) is a system that can index data to NHDPlus V2 catchments and offers a search service to discover indexed information. Data linked to the NLDI includes active NWIS stream gages , water quality portal sites , and outlets of HUC12 watersheds . The NLDI is a core product of the Internet of Water and is being developed as an open source project. .
In this blog post, we introduce the basic functions of the NLDI and show how to use it as a data discovery and access tool in R. The first section describes the operations available from the NLDI’s Web API . The second section shows how to map NLDI data and how to use the NLDI to discover data to be accessed with the dataRetrieval package.
Below, text highlighting is used in six ways:
- The names of API parameters such as
{featureSource} - Example values of API parameters such as:
USGS-08279500 - API operation names: navigation and basin
- API request names such as: getDataSources
- R functions such as: read_sf
- Other specific strings such as “siteNumber”
The NLDI Web API
The NLDI’s Web API follows a loosely RESTful design and is documented with swagger documentation which can be found here. Every request to get data from the NLDI starts from a given network linked feature.
Feature Sources
Available network linked feature sources ({featureSource}s) can be
found from the getDataSources
request.
(hint: Click the “try it out” button on the swagger page!) These are the
collections of network linked features the NLDI knows about. Think of
them as watershed outlets that can be used as a starting point. For this
demo, we’ll use NWIS Stream Gages as the {featureSource}. As a note
for later, {featureSource} here is the same as the {dataSource}
described below.
Feature IDs
All {featureID}s that can be accessed from a given {featureSource}
can be accessed with the
getFeatures
request. No filtering options are available. For this demo, we use a
well known NWIS
Streamgage
on the Rio Grande at Embudo
NM
as our starting point.

First USGS Stream Gage Rio Grande at Embudo
Indexed Features
We can use the
getRegisteredFeature
request to see this feature. Enter nwissite and USGS-08279500 in the
{featureSource} and {featureID}, respectively, in the swagger demo
page.
You can also see this in your browser at this url:
https://api.water.usgs.gov/nldi/linked-data/nwissite/USGS-08279500
The response contains the location of the feature, is in
geojson
, and looks like:
{
"type": "FeatureCollection",
"features": [
{
"type": "Feature",
"geometry": {
"type": "Point",
"coordinates": [
-105.9639722,
36.20555556
]
},
"properties": {
"source": "nwissite",
"sourceName": "NWIS Sites",
"identifier": "USGS-08279500",
"name": "RIO GRANDE AT EMBUDO, NM",
"uri": "https://waterdata.usgs.gov/nwis/inventory?agency_code=USGS&site_no=08279500",
"comid": "17864756",
"navigation": "https://api.water.usgs.gov/nldi/linked-data/nwissite/USGS-08279500/navigation"
}
}
]
}

Navigation
The navigation property of the returned feature is a url for the getNavigationTypes request. This request provides four navigation options as shown below. Each of these URLs returns the NHDPlus flowlines for the navigation type.
{
"upstreamMain": "https://api.water.usgs.gov/nldi/linked-data/nwissite/USGS-08279500/navigation/UM",
"upstreamTributaries": "https://api.water.usgs.gov/nldi/linked-data/nwissite/USGS-08279500/navigation/UT",
"downstreamMain": "https://api.water.usgs.gov/nldi/linked-data/nwissite/USGS-08279500/navigation/DM",
"downstreamDiversions": "https://api.water.usgs.gov/nldi/linked-data/nwissite/USGS-08279500/navigation/DD"
}
Get Flowlines from Navigation
Each of the URLs found via the getNavigationTypes request is a
complete
getFlowlines
request. This request has a required input parameter, {distance} that
allows specification of a distance to navigate in km. So, for example,
we can use this to retrieve data 150km upstream mainstem from the NWIS
gage 08279500 with a request like:
https://api.water.usgs.gov/nldi/linked-data/nwissite/USGS-08279500/navigation/UM/flowlines?distance=150
(Read below for more about the “flowlines” url path parameter)

Notice that the flowline goes downstream of the gage because the NLDI is referenced to whole NHDPlus catchments, not to precise network locations.
Get Linked Data from Navigation
Now that we have a {featureSource} = nwissite, a {featureID} =
USGS-082795001, the navigate operation on the feature, and the
{navigationMode} = UM with {distance} = 150km, we can use the
getFeatures
request to discover features from any {featureSource} which, in the
context of a getFeatures request, is called a {dataSource} – note
that above we used in flowlines {dataSource}. Setting the
{dataSource} = nwissite, we can see if there are any active NWIS
streamgages 150km upstream on the main stem with a request that looks
like:
https://api.water.usgs.gov/nldi/linked-data/nwissite/USGS-08279500/navigation/UM/nwissite?distance=150
Note that we could enter wqp in place of nwissite after UM here to
get water quality portal
sites
instead of NWIS sites. An example of this is shown later in this post.

Note: Click the black NWIS gage points to see a pop up and link!
Get the Upstream Basin Boundary
So far, we’ve covered four parameters of the NLDI Web API. The two base
parameters, {featureSource} and {featureID}, and two that apply to
the navigate option, {navigationMode} and {distance}. In
addition to the navigate option, the NLDI offers a basin
option for any {featureSource}/{featureID}. The
getBasin
operation doesn’t require any additional parameters, so a request to get
the basin for our stream gage looks like:
https://api.water.usgs.gov/nldi/linked-data/nwissite/USGS-08279500/basin
.

NLDI API Summary
There are a few other options available from the NLDI that are not covered here. One, that is coming soon, will make catchment (local incremental NHDPlus catchment) and basin (upstream accumulation) landscape characteristics available. This functionality and data is available but is preliminary and subject to change.
Bringing together all the operations summarized above, we can get:
- The NWIS site: https://api.water.usgs.gov/nldi/linked-data/nwissite/USGS-08279500
- The basin upstream of the site: https://api.water.usgs.gov/nldi/linked-data/nwissite/USGS-08279500/basin
- All upstream with tributaries flowlines: https://api.water.usgs.gov/nldi/linked-data/nwissite/USGS-08279500/navigation/UT/flowlines?distance=999
- The upstream mainstem flowlines: https://api.water.usgs.gov/nldi/linked-data/nwissite/USGS-08279500/navigation/UM/flowlines?distance=999
- The downstream mainstem flowlines: https://api.water.usgs.gov/nldi/linked-data/nwissite/USGS-08279500/navigation/DM/flowlines?distance=999
- The water quality observation sites in upstream catchments:
https://api.water.usgs.gov/nldi/linked-data/nwissite/USGS-08279500/navigation/UT/wqp?distance=999 - The water quality observations in downstream catchments:
https://api.water.usgs.gov/nldi/linked-data/nwissite/USGS-08279500/navigation/DM/wqp?distance=999
For QGIS users, you can use the NLDI URLs directly in the “Add Vector Layer” dialogue. The following two screenshots were rendered by loading the data into QGIS, turning on a base map with the OpenLayers Plugin, and applying a little styling to the NLDI layers. No local files needed!

Upstream Navigation Results

Downstream Navigation Results
Screenshots of NLDI data loaded into QGIS.
Using the NLDI in R.
Note: since this blog post was written, NLDI clients have been added to dataRetrieval and nhdplusTools in R and hyriver in python. The following is still a useful summary of how the NLDI API works via a scripting language.
First, we get all our URLs into a list and use read_sf from the sf package to download and read all the data into spatial data types. The end of this code block creates html for popup text with a web link tag that we’ll use later.
nldiURLs <- list(site_data = "https://api.water.usgs.gov/nldi/linked-data/nwissite/USGS-08279500",
basin_boundary = "https://api.water.usgs.gov/nldi/linked-data/nwissite/USGS-08279500/basin",
UT = "https://api.water.usgs.gov/nldi/linked-data/nwissite/USGS-08279500/navigation/UT/flowlines?distance=999",
UM = "https://api.water.usgs.gov/nldi/linked-data/nwissite/USGS-08279500/navigation/UM/flowlines?distance=999",
DM = "https://api.water.usgs.gov/nldi/linked-data/nwissite/USGS-08279500/navigation/DM/flowlines?distance=999",
UTwqp = "https://api.water.usgs.gov/nldi/linked-data/nwissite/USGS-08279500/navigation/UT/wqp?distance=999",
DMwqp = "https://api.water.usgs.gov/nldi/linked-data/nwissite/USGS-08279500/navigation/DM/wqp?distance=999")
nldi_data <- list()
for(n in names(nldiURLs)) {
nldi_data[n] <- list(sf::read_sf(nldiURLs[n][[1]]))
print(paste(n, "is of class", class(nldi_data[[n]]), "and has", nrow(nldi_data[[n]]), "features"))
}
## [1] "site_data is of class sf and has 1 features"
## [2] "site_data is of class tbl_df and has 1 features"
## [3] "site_data is of class tbl and has 1 features"
## [4] "site_data is of class data.frame and has 1 features"
## [1] "basin_boundary is of class sf and has 1 features"
## [2] "basin_boundary is of class tbl_df and has 1 features"
## [3] "basin_boundary is of class tbl and has 1 features"
## [4] "basin_boundary is of class data.frame and has 1 features"
## [1] "UT is of class sf and has 3371 features"
## [2] "UT is of class tbl_df and has 3371 features"
## [3] "UT is of class tbl and has 3371 features"
## [4] "UT is of class data.frame and has 3371 features"
## [1] "UM is of class sf and has 184 features"
## [2] "UM is of class tbl_df and has 184 features"
## [3] "UM is of class tbl and has 184 features"
## [4] "UM is of class data.frame and has 184 features"
## [1] "DM is of class sf and has 508 features"
## [2] "DM is of class tbl_df and has 508 features"
## [3] "DM is of class tbl and has 508 features"
## [4] "DM is of class data.frame and has 508 features"
## [1] "UTwqp is of class sf and has 1288 features"
## [2] "UTwqp is of class tbl_df and has 1288 features"
## [3] "UTwqp is of class tbl and has 1288 features"
## [4] "UTwqp is of class data.frame and has 1288 features"
## [1] "DMwqp is of class sf and has 1156 features"
## [2] "DMwqp is of class tbl_df and has 1156 features"
## [3] "DMwqp is of class tbl and has 1156 features"
## [4] "DMwqp is of class data.frame and has 1156 features"
UTwqp_html <- paste('<a href="',
nldi_data$UTwqp$uri,
'" target="_blank">',
nldi_data$UTwqp$name,
'</a>')
DMwqp_html <- paste('<a href="',
nldi_data$DMwqp$uri,
'" target="_blank">',
nldi_data$DMwqp$name,
'</a>')
Now that we have all our data, we can use leaflet functions to add the data to a map. First, let’s map just the upstream data. Note that the order we add them determines the order the layers are drawn. You can zoom in on the map shown and click the water quality sites to get a popup containing a link to the site’s landing page.
The get_base_map() function is described
here
and can be seen in
the source code of this blog
post.
map <- get_base_map()
map <- leaflet::addPolygons(map,
data=nldi_data$basin_boundary,
color = "black",
fill = FALSE,
weight = 1,
opacity = 1)
map <- leaflet::addPolylines(map,
data = nldi_data$UT,
color = "blue",
weight = 1,
opacity = 1)
map <- leaflet::addPolylines(map,
data = nldi_data$UM,
color = "blue",
weight = 3,
opacity = 0.5)
map <- leaflet::addCircleMarkers(map = map,
data = nldi_data$UTwqp,
radius = 1,
color = "black",
opacity = .5,
fill = FALSE,
popup = UTwqp_html)
map <- leaflet::addCircleMarkers(map,
data = nldi_data$site_data,
radius = 5,
color = "red")

To complete the picture, we can add the downstream main stem and water quality sites. Now we have an interactive map of all the upstream tributaries, water quality sites, basin boundary, the entire main stem, and water quality sites downstream.
map <- leaflet::addPolylines(map,
data = nldi_data$DM,
color = "blue",
weight = 3,
opacity = 0.5)
map <- leaflet::addCircleMarkers(map = map,
data = nldi_data$DMwqp,
radius = 1,
color = "black",
opacity = .5,
fill = FALSE,
popup = DMwqp_html)

This final map illustrates a very important detail about the NLDI if you zoom in on the downstream main stem. Notice that the sites are not all on the main stem flowpath. When the system indexes data sources that aren’t already indexed to particular reachcodes and measures along those reaches, it links sites (points) by looking at what local catchment polygon the site is in. This means that sites found through navigation may not be on the main flowpath of a catchment. In the future, we hope to improve the system such that it would know if indexed data are on or off the main flowpath of a catchment, but for now users need to be aware of this limitation.
Using the NLDI to discover linked observations data.
The two sources of linked data shown above, nwissite and wqp, are
both queryable from the dataRetrieval
package.
The
“siteNumber” input of the dataRetrieval functions that start with
“readNWIS” can be found by removing “USGS-” from the “identifier”
attribute of features found using nwissite as the {dataSource}
input. The following code shows the NLDI identifiers and how to use them
with the dataRetrieval function readNWISdv.
nwis_gage_url <- "https://api.water.usgs.gov/nldi/linked-data/nwissite/USGS-08279500/navigation/UM/nwissite?distance=150"
nwis_gages <- sf::read_sf(nwis_gage_url)
nwis_ids <- as.character(nwis_gages$identifier)
print(paste("The NLDI ID for:", nwis_gages$name, "is", nwis_ids))
## [1] "The NLDI ID for: RIO GRANDE NEAR LOBATOS, CO is USGS-08251500"
## [2] "The NLDI ID for: RIO GRANDE NEAR CERRO, NM is USGS-08263500"
## [3] "The NLDI ID for: RIO GRANDE BLW TAOS JUNCTION BRIDGE NEAR TAOS, NM is USGS-08276500"
## [4] "The NLDI ID for: RIO GRANDE AT EMBUDO, NM is USGS-08279500"
nwis_ids <- gsub(pattern = "USGS-", replacement = "", nwis_ids)
print(paste(nwis_gages$name, "has id", nwis_ids))
## [1] "RIO GRANDE NEAR LOBATOS, CO has id 08251500"
## [2] "RIO GRANDE NEAR CERRO, NM has id 08263500"
## [3] "RIO GRANDE BLW TAOS JUNCTION BRIDGE NEAR TAOS, NM has id 08276500"
## [4] "RIO GRANDE AT EMBUDO, NM has id 08279500"
# Now we can use these IDs with dataRetrieval.
dv_data <- dataRetrieval::readNWISdv(siteNumber = nwis_ids[1], parameterCd = '00060')
plot(dv_data$Date, dv_data$X_00060_00003,
main = paste("Daily Streamflow for", nwis_gages$name[1]), xlab = "", ylab = "Daily Streamflow (CFS)")
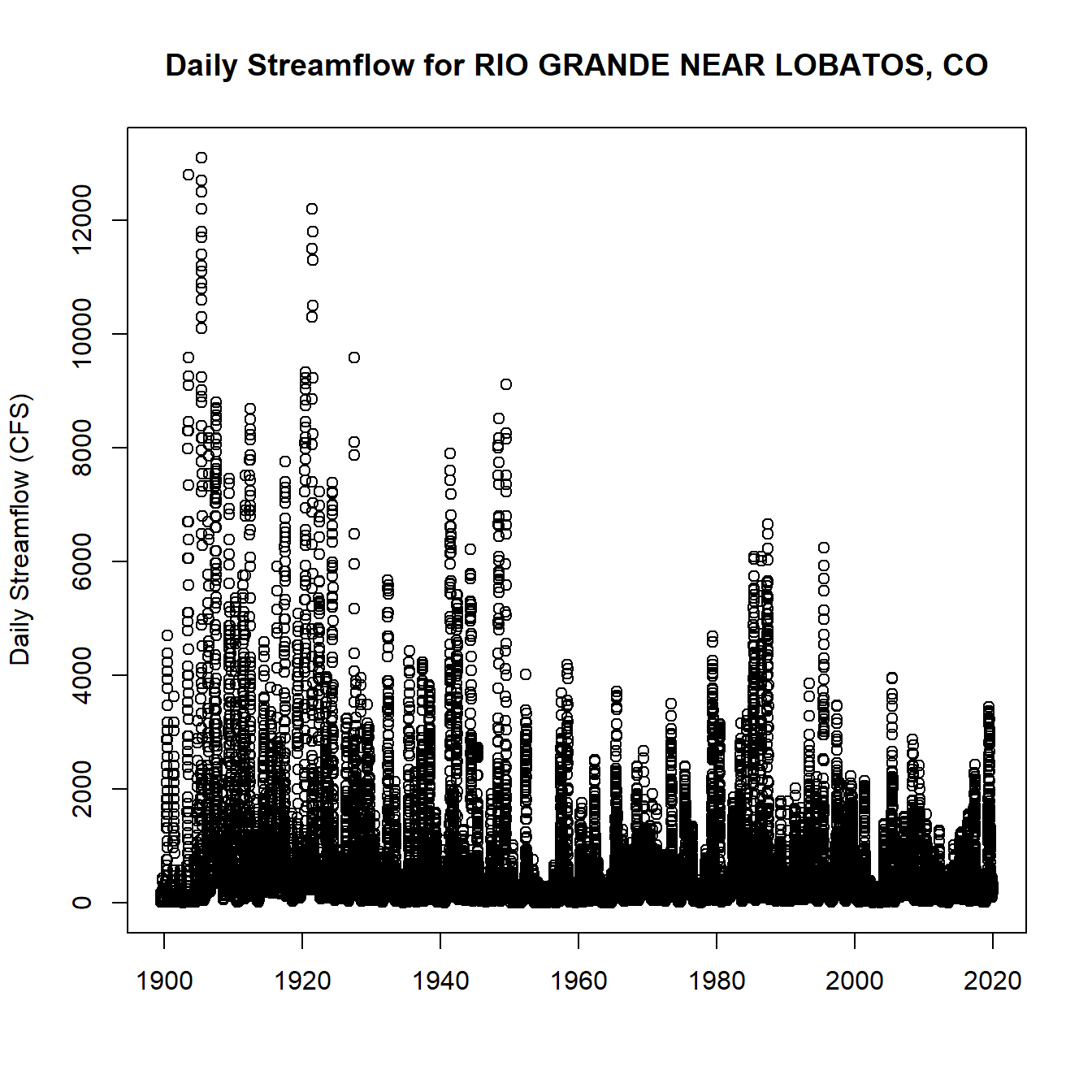
Daily Streamflow Plot
Similarly, we can use identifiers returned using wqp as the
{dataSource} with the readWQPqw function. In this case, the
identifiers can be used without modification as shown below. Note that
the NLDI query for downstream mainstem found 2756 sites and upstream
tributaries found 1908 sites. The query below gets data from just one!
The NLDI is used as a spatial pre-filter in the Water Quality Portal
user interface
, which has a
rich set of filter options in addition to network navigation.
wqp_site <- list(names = as.character(nldi_data$DMwqp$name),
ids = as.character(nldi_data$DMwqp$identifier))
print(paste(wqp_site$names[1:10], "has id", wqp_site$ids[1:10]))
## [1] "JL-49-05-622 has id USGS-315655106231501"
## [2] "Rio Grande at Highway 74 near San Juan Pueblo has id 21NMEX-28RGRAND572.8"
## [3] "Los Lunas WWTP effluent - NM0020303-A has id 21NMEX-NM0020303-A"
## [4] "RIO GRANDE AT CORCHESNE BRIDGE- 42RGrand002.7 has id 21NMEX_WQX-42RGrand002.7"
## [5] "New Mexico 147 Bridge has id PUEBLOISLETA-NM147"
## [6] "Confluence has id PUEBLO_SANTAANA-RG2"
## [7] "Rio Grande above Rio Rancho WWTP #3 outfall. has id SANDIAWQ_WQX-RG005"
## [8] "RG014 Rio Grande, straight west of the Fire Bridge. has id SANDIAWQ_WQX-RG014"
## [9] "Wetland pond TLIP pond-southside has id SANDIAWQ_WQX-WETPONDTLIP-SO"
## [10] "On Tonque Arroyo, 100ft above the River has id SANFELIPE_WQX-Site5"
wqp_data <- dataRetrieval::readWQPqw(siteNumbers = wqp_site$ids[1:10], parameterCd = "")
print(paste0("Got ", ncol(wqp_data), " samples beween ", min(wqp_data$ActivityStartDate), " and ", max(wqp_data$ActivityStartDate), " for characteristics: ", paste(unique(wqp_data$CharacteristicName), collapse = ", ")))
## [1] "Got 65 samples beween 1990-08-24 and 2020-09-18 for characteristics: pH, Dissolved oxygen (DO), Turbidity, Temperature, water, Barometric pressure, Specific conductance, Acidity, hydrogen ion (H+), Oxygen, Carbon dioxide, Carbonate, Bicarbonate, Organic carbon, Hardness, Ca, Mg, Hardness, non-carbonate, Calcium, Magnesium, Sodium, Sodium adsorption ratio [(Na)/(sq root of 1/2 Ca + Mg)], Sodium, percent total cations, Potassium, Chloride, Sulfate, Fluoride, Silica, Boron, MBAS, Alkalinity, Total dissolved solids, Iodide, Bromide, Carbon-13/Carbon-12 ratio, Deuterium/Hydrogen ratio, Oxygen-18/Oxygen-16 ratio, Nitrogen, mixed forms (NH3), (NH4), organic, (NO2) and (NO3), Organic Nitrogen, Ammonia and ammonium, Nitrite, Nitrate, Kjeldahl nitrogen, Inorganic nitrogen (nitrate and nitrite), Orthophosphate, Phosphorus, Aluminum, Dichlorobromomethane, Carbon tetrachloride, 1,2-Dichloroethane, Tribromomethane, Chlorodibromomethane, Chloroform, Toluene, Benzene, Chlorobenzene, Chloroethane, Ethylbenzene, Methyl bromide, Chloromethane, Methylene chloride, Tetrachloroethylene, CFC-11, 1,1-Dichloroethane, 1,1-Dichloroethylene, 1,1,1-Trichloroethane, 1,1,2-Trichloroethane, 1,1,2,2-Tetrachloroethane, o-Dichlorobenzene, 1,2-Dichloropropane, trans-1,2-Dichloroethylene, 1,3-Dichloropropene, m-Dichlorobenzene, p-Dichlorobenzene, 2-Chloroethyl vinyl ether, CFC-12, trans-1,3-Dichloropropene, cis-1,3-Dichloropropene, Vinyl chloride, Trichloroethylene, Styrene, Ethylene dibromide, Xylene, Trihalomethanes, Acenaphthylene, Acenaphthene, Anthracene, Benzo(b)fluoranthene, Benzo[k]fluoranthene, Benzo[a]pyrene, Bis(2-chloroethyl) ether, Bis(2-chloroethoxy)methane, Bis(2-chloroisopropyl) ether, Butyl benzyl phthalate, Chrysene, Diethyl phthalate, Dimethyl phthalate, Fluoranthene, Fluorene, Hexachlorocyclopentadiene, Hexachloroethane, Indeno[1,2,3-cd]pyrene, Isophorone, N-Nitrosodi-n-propylamine, N-Nitrosodiphenylamine, N-Nitrosodimethylamine, Nitrobenzene, p-Chloro-m-cresol, Phenanthrene, Pyrene, Benzo[g,h,i]perylene, Benz[a]anthracene, 1,2,4-Trichlorobenzene, Dibenz[a,h]anthracene, 2-Chloronaphthalene, o-Chlorophenol, o-Nitrophenol, Di-n-octyl phthalate, 2,4-Dichlorophenol, 2,4-Dimethylphenol, 2,4-Dinitrotoluene, 2,4-Dinitrophenol, 2,4,6-Trichlorophenol, 2,6-Dinitrotoluene, BDE-003, p-Chlorophenyl phenyl ether, p-Nitrophenol, 4,6-Dinitro-o-cresol, Phenol, Naphthalene, Pentachlorophenol, Di(2-ethoxylhexyl) phthalate, Dibutyl phthalate, Hexachlorobenzene, Hexachlorobutadiene, Inorganic nitrogen (nitrate and nitrite) as N, Sulfate as SO4, Alkalinity, total, Dissolved oxygen saturation, Total solids, Ammonia, 2,2',3,4',5,6'-Hexachlorobiphenyl, PCB-90/101/113, 2,3,3',4,6-Pentachlorobiphenyl, 2,2',4,6,6'-Pentachlorobiphenyl, 2,2',3,4,5',6-Hexachlorobiphenyl, 2,2',3,4',6,6'-Hexachlorobiphenyl, 2,2',3,5,6,6'-Hexachlorobiphenyl, 3,4,4',5-Tetrachlorobiphenyl, 3,3',4,5,5'-Pentachlorobiphenyl, 2,2',3,4',5,6,6'-Heptachlorobiphenyl, 2,3',5,5'-Tetrachlorobiphenyl, PCB-129/138/160/163, PCB-93/95/98/100/102, PCB-107/124, PCB-40/41/71, PCB-21/33, PCB-20/28, PCB-139/140, 2,2',3,5,5'-Pentachlorobiphenyl, 2,2'-Dichlorobiphenyl, 2,2',3,3',4,5'-Hexachlorobiphenyl, PCB-197/200, PCB-83/99, PCB-110/115, PCB-86/87/97/108/119/125, 2,2',3,3',4,4',5,6,6'-Nonachlorobiphenyl, 2,2',3,5,6'-Pentachlorobiphenyl, 3,3',4,5'-Tetrachlorobiphenyl, Hexachlorobiphenyl, Trichlorobiphenyl, 3,4',5-Trichlorobiphenyl, 2,2',3,6'-Tetrachlorobiphenyl, 2,2',3,3',4,4',5,6-Octachlorobiphenyl, 2,3,4,4',5-Pentachlorobiphenyl, 2,2',3,3',4,4',5,5',6-Nonachlorobiphenyl, 3,3',4,5-Tetrachlorobiphenyl, 2,2',6-Trichlorobiphenyl, 2,3,4',6-Tetrachlorobiphenyl, PCB-12/13, 2,3,3',4-Tetrachlorobiphenyl, 2,2',3,3',6-Pentachlorobiphenyl, 2,3,3',4,5,5'-Hexachlorobiphenyl, 2,2',3,4',5,5'-Hexachlorobiphenyl, 3-Chlorobiphenyl, PCB-128/166, 4,4'-Dichlorobiphenyl, Monochlorobiphenyl, PCB-50/53, 2,2',4,4',6,6'-Hexachlorobiphenyl, 3,3',4-Trichlorobiphenyl, Polychlorinated biphenyls, 2,3,3',5,5',6-Hexachlorobiphenyl, 2,3,3',5,6-Pentachlorobiphenyl, 2,2',3,3',5,5'-Hexachlorobiphenyl, 2,2',3,4,5,6-Hexachlorobiphenyl, 2,3,3',4,5',6-Hexachlorobiphenyl, 2,2',3,3',5,5',6,6'-Octachlorobiphenyl, 2,2',3,3',4,6'-Hexachlorobiphenyl, Octachlorobiphenyl, PCB-183/185, 4-Chlorobiphenyl, 2,2',3,4,4',5,5',6-Octachlorobiphenyl, 2,3,4',5-Tetrachlorobiphenyl, 3,3'-Dichlorobiphenyl, 2,3',4,5'-Tetrachlorobiphenyl, 2,3,5-Trichlorobiphenyl, 2,3,3',5-Tetrachlorobiphenyl, 2,2',4-Trichlorobiphenyl, Pentachlorobiphenyl, 3,3',4,4'-Tetrachlorobiphenyl, 2,2',3,4,4',5,6-Heptachlorobiphenyl, 2,2',3,3',4,6-Hexachlorobiphenyl, 3,5-Dichlorobiphenyl, 2,2',3,3',4,5,5',6,6'-Nonachlorobiphenyl, 2,3',5'-Trichlorobiphenyl, 2,2',3,6,6'-Pentachlorobiphenyl, 3,3',5,5'-Tetrachlorobiphenyl, 2,3,3',4,4',5,5'-Heptachlorobiphenyl, PCB-198/199, PCB-26/29, 3,3',5-Trichlorobiphenyl, 2,3,3',4'-Tetrachlorobiphenyl, 2,4,6-Trichlorobiphenyl, 2,3',4,5-Tetrachlorobiphenyl, 2,2',3,3',4-Pentachlorobiphenyl, 2,2',3,3',4,5',6-Heptachlorobiphenyl, 2,3,3',4',5'-Pentachlorobiphenyl, 2,3,3',5,5'-Pentachlorobiphenyl, 2,3',4,5',6-Pentachlorobiphenyl, 2,2',3,4,6,6'-Hexachlorobiphenyl, 2,2',3,4,4',6,6'-Heptachlorobiphenyl, 2,2',3,4,4',5,6'-Heptachlorobiphenyl, PCB-85/116/117, 2,2',3,4'-Tetrachlorobiphenyl, PCB-45/51, 2,4-Dichlorobiphenyl, 2,3,6-Trichlorobiphenyl, 2,2',3,4,4',5,6,6'-Octachlorobiphenyl, 2,2',3,3',4,4',5,6'-Octachlorobiphenyl, 2,2',3-Trichlorobiphenyl, 2,2',3,3',4,6,6'-Heptachlorobiphenyl, 2,2',4,5-Tetrachlorobiphenyl, PCB-18/30, PCB-61/70/74/76, PCB-135/151/154, PCB-44/47/65, 2,3,4'-Trichlorobiphenyl, 2,2',3,4,6'-Pentachlorobiphenyl, 2,3,4,4'-Tetrachlorobiphenyl, 2,3',4-Trichlorobiphenyl, 2,2',3,3',4,5,5'-Heptachlorobiphenyl, 2,2',3,3',5,5',6-Heptachlorobiphenyl, Nonachlorobiphenyl, Dichlorobiphenyl, 2,3,3',4,4'-Pentachlorobiphenyl, 2-Chlorobiphenyl, 2,2',3,3',4,4',5-Heptachlorobiphenyl, 3,3',4,4',5-Pentachlorobiphenyl, 2,3,3',4,5-Pentachlorobiphenyl, 2,3,3',4,4',6-Hexachlorobiphenyl, 2,2',3,3',4,4',5,5'-Octachlorobiphenyl, 2,2',3,3',4,5',6'-Heptachlorobiphenyl, 2,2',3,3',4,5',6,6'-Octachlorobiphenyl, 2,4'-Dichlorobiphenyl, Total nonfecal coliform, 2,3,3',4,4',5,5',6-Octachlorobiphenyl, Heptachlorobiphenyl, 2,2',3,3',4,5,6'-Heptachlorobiphenyl, 2,2',5,5'-Tetrachlorobiphenyl, 2,3',4,4',5-Pentachlorobiphenyl, 2,5-Dichlorobiphenyl, 2,2',3,3',5,6,6'-Heptachlorobiphenyl, 2,6-Dichlorobiphenyl, 2,2',3,3',6,6'-Hexachlorobiphenyl, 2,2',3,4,5,5'-Hexachlorobiphenyl, 3,4,5-Trichlorobiphenyl, 3,4,4'-Trichlorobiphenyl, 2,3,3',4',5,5'-Hexachlorobiphenyl, 2,3,3',4',5',6-Hexachlorobiphenyl, 2,3',4,5,5'-Pentachlorobiphenyl, 2,2',4,5',6-Pentachlorobiphenyl, 2,2',3,4,5,6,6'-Heptachlorobiphenyl, PCB-59/62/75, PCB-171/173, 2,3,3',4,4',5',6-Heptachlorobiphenyl, PCB-147/149, PCB-49/69, 2,3'-Dichlorobiphenyl, PCB-156/157, 2,2',3,4,4',5-Hexachlorobiphenyl, 2,4',5-Trichlorobiphenyl, PCB-134/143, PCB-88/91, Tetrachlorobiphenyl, 3,3',4,4',5,5'-Hexachlorobiphenyl, PCB-153/168, 2,3',4,4',5'-Pentachlorobiphenyl, 2,3',4,4',5,5'-Hexachlorobiphenyl, 2,2',3,4',5,5',6-Heptachlorobiphenyl, 2,2',3,5-Tetrachlorobiphenyl, 2,2',6,6'-Tetrachlorobiphenyl, Decachlorobiphenyl, 2,3',4,4'-Tetrachlorobiphenyl, 2,3,3',4,5,5',6-Heptachlorobiphenyl, 2,3',5',6-Tetrachlorobiphenyl, 2,3,3',4,4',5,6-Heptachlorobiphenyl, 2,3',6-Trichlorobiphenyl, 2,3,3',5'-Tetrachlorobiphenyl, 2,3-Dichlorobiphenyl, PCB-180/193, Nitrogen, Escherichia coli, Triadimefon, Endrin, Propachlor, 3,5-Dichlorobenzoic acid, p,p'-DDD, 1-Methylnaphthalene, 2,4-D, Endrin aldehyde, .alpha.-Endosulfan, Lindane, Methoxychlor, Oxidation reduction potential (ORP), Conductivity, .beta.-Endosulfan, Benzene Hexachloride, Delta (BHC), Trifluralin, Aroclor 1232, Heptachlor, Endosulfan sulfate, Benzene Hexachloride, Beta (BHC), Silvex, p,p'-DDE, Heptachlor epoxide, Salinity, Picloram, Metribuzin, Endrin ketone, Cyanazine, Vanadium, Dieldrin, Depth, trans-Nonachlor, Metolachlor, Benzidine, Dichlorprop, Aroclor 1016, Aroclor 1221, Cobalt, Diazinon, Pendimethalin, Benzene Hexachloride, Alpha (BHC), Chlorthal-dimethyl, Aroclor 1260, p,p'-DDT, Dinoseb, 2,4,5-T, Aroclor 1254, Arsenic, Acifluorfen, Aroclor 1242, Cyanide, Benzo[ghi]perylene, Molybdenum, Malathion, Beryllium, Chloramben, 2-Methylnaphthalene, Parathion, Butachlor, Thallium, Maximum total trihalomethane potential, Chlorpyrifos, Selenium, Chlordane, S-Ethyl dipropylthiocarbamate, Prometon, trans-Chlordane, Aroclor 1248, Barium, Di(2-ethylhexyl) adipate, Bentazon, Toxaphene, Acrolein, Terbacil, Dicamba, Cadmium, 1,3-Dichlorobenzene, 2,4-DB, Di(2-ethylhexyl) phthalate, Simazine, Bromacil, Atrazine, Depth, Secchi disk depth, Dalapon, cis-Chlordane, Permethrin, Velocity-discharge, Temperature, air, Acrylonitrile, Lithium, Phosphate-phosphorus, Aldrin, cis-1,2-Dichloroethylene, Alachlor, Chromium, Copper, Nitrate + Nitrite, Antimony, Total suspended solids, Beta particle, Wind velocity, Wind direction (direction from, expressed 0-360 deg), Relative humidity, Precipitation 24hr prior to monitoring event amount, Zinc, Lead, Total Nitrogen, mixed forms, Ammonia-nitrogen, Velocity - stream, p-Chlorotoluene, Nickel, Iron, Biochemical oxygen demand, standard conditions, Manganese, Sulfotep, Acetone, Chloroprene, o-Chlorotoluene, .beta.-Hexachlorocyclohexane, Hardness, carbonate, Silver, Total Coliform, Fecal Coliform, Alpha particle, Acetonitrile, Methyl methacrylate, Methacrylonitrile, Mercury, p-Dinitrobenzene, Aniline, Chromium(VI), 2,4,5-Trichlorophenol, 1,2-Dibromo-3-chloropropane, 1,1,1,2-Tetrachloroethane, Vinyl acetate, 1,3,5-Trimethylbenzene, Flow, Dibenzofuran, Ronnel, Gross beta radioactivity, (Cesium-137 ref std), Gross alpha radioactivity, (nat-Uranium ref std), 2-Hexanone, Bis(2-chloro-1-methylethyl) ether, Carbon disulfide, Prometryn, .delta.-Hexachlorocyclohexane, 1,2,4-Trimethylbenzene, m-Dinitrobenzene, 1,1-Dichloropropene, Carbazole, Bromobenzene, n-Propylbenzene, o-Xylene, Tetrahydrofuran, Methyl ethyl ketone, 1,4-Dichloro-2-butene, n-Butylbenzene, m-Nitroaniline, 2,3,5,6-Tetrachlorophenol, tert-Butylbenzene, trans-1,4-Dichloro-2-butene, 1,4-Dioxane, Ethyl methacrylate, Methyl iodide, 1,2,3-Trichlorobenzene, Propanenitrile, Dibromomethane, Benzyl alcohol, o-Nitroaniline, m-Cymene, Allyl chloride, Gross beta radioactivity, (Strontium-Yttrium-90 ref std), Pentachloroethane, 1,2,3-Trichloropropane, cis-Nonachlor, m,p-Xylene, 2,2-Dichloropropane, 3,3'-Dichlorobenzidine, 1,3-Dichloropropane, .alpha.-Hexachlorocyclohexane, p-Nitroaniline, 2,3,4,6-Tetrachlorophenol, Isobutanol, Cumene, Pyridine, Azobenzene, Halon 1011, Methyl isobutyl ketone, O-Dinitrobenzene, p-Chloroaniline, Gross alpha radioactivity, (Americium-241 ref std)"
Summary
In this blog post, we summarized the NLDI’s Web API through links to the
system’s
Swagger
documentation. The primary API parameters, {featureSource} and
{featureID}, were described. Two functions that operate with any
{featureID}, navigation (and it’s optional {distance}
parameter) and basin were demonstrated. The the navigation
function’s {dataSource} parameter, which can be any {featureSource},
was shown by retrieving NWIS (nwissite) and WQP (wqp) sites upstream
and downstream of an NWIS site.
The post finishes by showing how to use sites found with the NLDI to download data from the National Water Information System and Water Quality portal. The potential for extending this use of the NLDI is vast. As more feature/data sources are indexed and the system evolves, the NLDI should serve as a major new discovery service for many sources of observed and modeled data.
The NLDI is an exciting new service that is being implemented in an incremental and agile development process. Given that, the API will expand and new versions of the API may have somewhat different design. The intention is to keep the Web API described here the same, only changing it by introducing a version identifier as an API parameter. If you found this useful and plan on using the NLDI as a dependency in a project of application, we would greatly appreciate hearing about your use case and can answer any questions you have while implementing your application.
Please email dblodgett@usgs.gov with questions and feedback.
Categories:
Related Posts
Network Linked Data Index (NLDI) Update and Client Applications
September 30, 2020
In August 2020, the Hydro Network Linked Data Index (NLDI) was updated with new functionality and some changes to the existing Web Application Programming Interface (API). This post summarizes these changes and demonstrates Python and R clients available to work with the functionality.
The Network Linked Data Index (NLDI) Geoprocessing with OGC API Processes
April 12, 2022
The Network Linked Data Index (NLDI) is a search engine that indexes data to the flowpaths and/or catchments of a river network and provides discovery services based on position in that network. Like a search engine, it can cache and index new data. It also offers some convenient data services like basin boundaries and accumulated catchment characteristics.
Cloud-optimized USGS time series data
January 8, 2020
Intro
The scalability, flexibility, and configurabilty of the commercial cloud provide new and exciting possibilities for many applications including earth science research. As with many organizations, the US Geological Survey (USGS) is exploring the use and advantages of the cloud for performing their science. The full benefit of the cloud, however, can only be realized if the data that will be used is in a cloud-friendly format.
A map that glows with the vocabulary of water
February 27, 2026
English is the official language and authoritative version of all federal information.
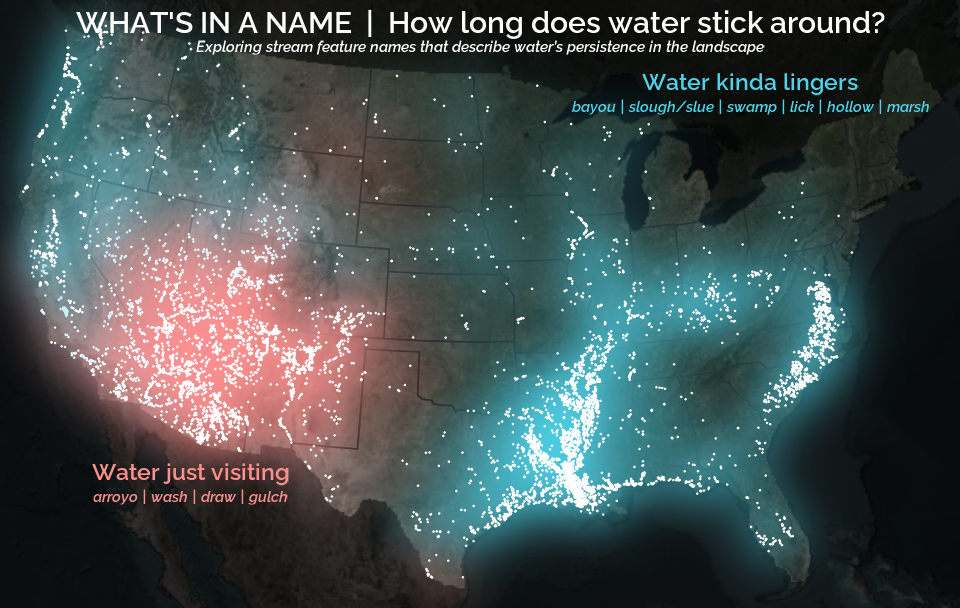
A map that glows with the vocabulary of water
What is your first impression of the map above? To me, it is the shimmer. Thousands of points of light, each one a stream or river, illuminating a darkened basemap. Look closely and a pattern emerges: the country’s waterways form a linguistic constellation. These points are classified not from population data or even explicitly by hydrology. They glow strictly according to the vocabulary used to name them and what can be implied about the hydrology of these streams based on their names.
Extracting the grammar of U.S. stream names
February 27, 2026
English is the official language and authoritative version of all federal information.
Extracting a stream’s feature
The names of streams (hydronyms ) contain, hidden within them, the power to show us the linguistic patterns within the United States. In the United States, stream names tend to follow a binomial structure: a specific name (“Moose,” “Columbia,” “Snake”) paired with a generic feature word (“creek,” “river,” “fork,” “bayou”). The specific portion is endlessly variable, but the generic part is surprisingly stable. In fact, if you look at stream names across the country, the diversity of generic terms is relatively small, but shaped by centuries of hydrologic realities, settlement history, and local tradition.

